matrixStats.benchmarks
varDiff() benchmarks on subsetted computation
This report benchmark the performance of varDiff() on subsetted computation.
Data type “integer”
Data
> rvector <- function(n, mode = c("logical", "double", "integer"), range = c(-100, +100), na_prob = 0) {
+ mode <- match.arg(mode)
+ if (mode == "logical") {
+ x <- sample(c(FALSE, TRUE), size = n, replace = TRUE)
+ } else {
+ x <- runif(n, min = range[1], max = range[2])
+ }
+ storage.mode(x) <- mode
+ if (na_prob > 0)
+ x[sample(n, size = na_prob * n)] <- NA
+ x
+ }
> rvectors <- function(scale = 10, seed = 1, ...) {
+ set.seed(seed)
+ data <- list()
+ data[[1]] <- rvector(n = scale * 100, ...)
+ data[[2]] <- rvector(n = scale * 1000, ...)
+ data[[3]] <- rvector(n = scale * 10000, ...)
+ data[[4]] <- rvector(n = scale * 1e+05, ...)
+ data[[5]] <- rvector(n = scale * 1e+06, ...)
+ names(data) <- sprintf("n = %d", sapply(data, FUN = length))
+ data
+ }
> data <- rvectors(mode = mode)
> data <- data[1:4]
Results
n = 1000 vector
All elements
> x <- data[["n = 1000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5362844 286.5 7916910 422.9 7916910 422.9
Vcells 11153947 85.1 39038428 297.9 94934136 724.3
> stats <- microbenchmark(varDiff_x_S = varDiff(x_S), `varDiff(x, idxs)` = varDiff(x, idxs = idxs),
+ `varDiff(x[idxs])` = varDiff(x[idxs]), unit = "ms")
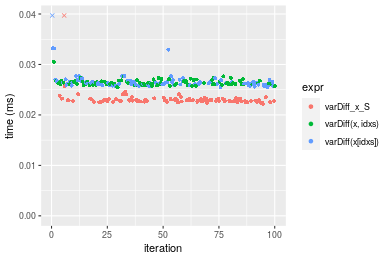
Table: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on integer+n = 1000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 0.022212 | 0.0226885 | 0.0236181 | 0.0228800 | 0.023157 | 0.087388 |
| 3 | varDiff(x[idxs]) | 0.025511 | 0.0260000 | 0.0290972 | 0.0262840 | 0.026625 | 0.281070 |
| 2 | varDiff(x, idxs) | 0.025429 | 0.0260535 | 0.0264321 | 0.0263085 | 0.026717 | 0.030500 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 3 | varDiff(x[idxs]) | 1.148523 | 1.145955 | 1.231988 | 1.148776 | 1.149760 | 3.2163455 |
| 2 | varDiff(x, idxs) | 1.144832 | 1.148313 | 1.119146 | 1.149847 | 1.153733 | 0.3490182 |
Figure: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on integer+n = 1000 data. Outliers are displayed as crosses. Times are in milliseconds.
n = 10000 vector
All elements
> x <- data[["n = 10000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5359912 286.3 7916910 422.9 7916910 422.9
Vcells 10899708 83.2 39038428 297.9 94934136 724.3
> stats <- microbenchmark(varDiff_x_S = varDiff(x_S), `varDiff(x, idxs)` = varDiff(x, idxs = idxs),
+ `varDiff(x[idxs])` = varDiff(x[idxs]), unit = "ms")
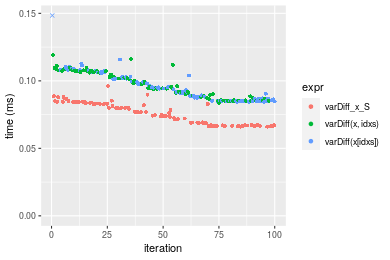
Table: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on integer+n = 10000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 0.066079 | 0.0671120 | 0.0757118 | 0.0753615 | 0.0830735 | 0.096079 |
| 3 | varDiff(x[idxs]) | 0.084400 | 0.0867175 | 0.0959805 | 0.0942560 | 0.1029010 | 0.181812 |
| 2 | varDiff(x, idxs) | 0.083807 | 0.0856580 | 0.0961852 | 0.0946705 | 0.1064495 | 0.119259 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 3 | varDiff(x[idxs]) | 1.277259 | 1.292131 | 1.267709 | 1.250718 | 1.238674 | 1.892318 |
| 2 | varDiff(x, idxs) | 1.268285 | 1.276344 | 1.270412 | 1.256218 | 1.281389 | 1.241260 |
Figure: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on integer+n = 10000 data. Outliers are displayed as crosses. Times are in milliseconds.
n = 100000 vector
All elements
> x <- data[["n = 100000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5359984 286.3 7916910 422.9 7916910 422.9
Vcells 10963268 83.7 39038428 297.9 94934136 724.3
> stats <- microbenchmark(varDiff_x_S = varDiff(x_S), `varDiff(x, idxs)` = varDiff(x, idxs = idxs),
+ `varDiff(x[idxs])` = varDiff(x[idxs]), unit = "ms")
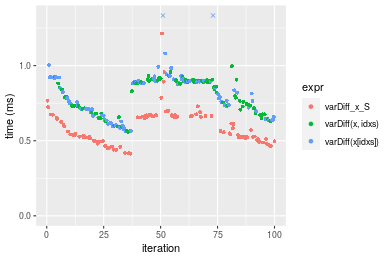
Table: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on integer+n = 100000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 0.413811 | 0.5045290 | 0.5857393 | 0.544012 | 0.6621690 | 1.213714 |
| 3 | varDiff(x[idxs]) | 0.561553 | 0.6705570 | 0.9228240 | 0.789193 | 0.8980275 | 13.785594 |
| 2 | varDiff(x, idxs) | 0.560413 | 0.7139785 | 0.7980298 | 0.835218 | 0.8945030 | 0.995830 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 3 | varDiff(x[idxs]) | 1.357028 | 1.329075 | 1.575486 | 1.450690 | 1.356191 | 11.3581898 |
| 2 | varDiff(x, idxs) | 1.354273 | 1.415139 | 1.362432 | 1.535293 | 1.350868 | 0.8204816 |
Figure: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on integer+n = 100000 data. Outliers are displayed as crosses. Times are in milliseconds.
n = 1000000 vector
All elements
> x <- data[["n = 1000000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5360056 286.3 7916910 422.9 7916910 422.9
Vcells 11593317 88.5 39038428 297.9 94934136 724.3
> stats <- microbenchmark(varDiff_x_S = varDiff(x_S), `varDiff(x, idxs)` = varDiff(x, idxs = idxs),
+ `varDiff(x[idxs])` = varDiff(x[idxs]), unit = "ms")
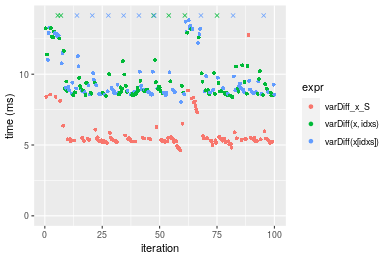
Table: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on integer+n = 1000000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 4.635450 | 5.266083 | 5.782645 | 5.402392 | 5.53224 | 12.76769 |
| 2 | varDiff(x, idxs) | 8.392257 | 8.700843 | 10.176747 | 8.977796 | 10.06822 | 36.72024 |
| 3 | varDiff(x[idxs]) | 8.040496 | 8.723145 | 10.411642 | 9.023121 | 11.38572 | 18.65386 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | varDiff(x, idxs) | 1.810451 | 1.652242 | 1.759878 | 1.661819 | 1.819917 | 2.876029 |
| 3 | varDiff(x[idxs]) | 1.734566 | 1.656477 | 1.800498 | 1.670209 | 2.058066 | 1.461021 |
Figure: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on integer+n = 1000000 data. Outliers are displayed as crosses. Times are in milliseconds.
Data type “double”
Data
> rvector <- function(n, mode = c("logical", "double", "integer"), range = c(-100, +100), na_prob = 0) {
+ mode <- match.arg(mode)
+ if (mode == "logical") {
+ x <- sample(c(FALSE, TRUE), size = n, replace = TRUE)
+ } else {
+ x <- runif(n, min = range[1], max = range[2])
+ }
+ storage.mode(x) <- mode
+ if (na_prob > 0)
+ x[sample(n, size = na_prob * n)] <- NA
+ x
+ }
> rvectors <- function(scale = 10, seed = 1, ...) {
+ set.seed(seed)
+ data <- list()
+ data[[1]] <- rvector(n = scale * 100, ...)
+ data[[2]] <- rvector(n = scale * 1000, ...)
+ data[[3]] <- rvector(n = scale * 10000, ...)
+ data[[4]] <- rvector(n = scale * 1e+05, ...)
+ data[[5]] <- rvector(n = scale * 1e+06, ...)
+ names(data) <- sprintf("n = %d", sapply(data, FUN = length))
+ data
+ }
> data <- rvectors(mode = mode)
> data <- data[1:4]
Results
n = 1000 vector
All elements
> x <- data[["n = 1000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5360131 286.3 7916910 422.9 7916910 422.9
Vcells 11450125 87.4 39038428 297.9 94934136 724.3
> stats <- microbenchmark(varDiff_x_S = varDiff(x_S), `varDiff(x, idxs)` = varDiff(x, idxs = idxs),
+ `varDiff(x[idxs])` = varDiff(x[idxs]), unit = "ms")
Table: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on double+n = 1000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 0.019873 | 0.0202375 | 0.0205991 | 0.0204165 | 0.0208305 | 0.022106 |
| 2 | varDiff(x, idxs) | 0.022824 | 0.0232165 | 0.0238168 | 0.0234755 | 0.0237265 | 0.040249 |
| 3 | varDiff(x[idxs]) | 0.022967 | 0.0233720 | 0.0242683 | 0.0236040 | 0.0238445 | 0.080263 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | varDiff(x, idxs) | 1.148493 | 1.147202 | 1.156203 | 1.149830 | 1.139027 | 1.820727 |
| 3 | varDiff(x[idxs]) | 1.155689 | 1.154886 | 1.178122 | 1.156124 | 1.144692 | 3.630824 |
Figure: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on double+n = 1000 data. Outliers are displayed as crosses. Times are in milliseconds.
n = 10000 vector
All elements
> x <- data[["n = 10000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5360200 286.3 7916910 422.9 7916910 422.9
Vcells 11459871 87.5 39038428 297.9 94934136 724.3
> stats <- microbenchmark(varDiff_x_S = varDiff(x_S), `varDiff(x, idxs)` = varDiff(x, idxs = idxs),
+ `varDiff(x[idxs])` = varDiff(x[idxs]), unit = "ms")
Table: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on double+n = 10000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 0.051286 | 0.0566970 | 0.0604725 | 0.0604575 | 0.0612570 | 0.119799 |
| 2 | varDiff(x, idxs) | 0.070962 | 0.0764785 | 0.0825251 | 0.0808835 | 0.0832660 | 0.159821 |
| 3 | varDiff(x[idxs]) | 0.068973 | 0.0763860 | 0.0826795 | 0.0809315 | 0.0841145 | 0.157204 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | varDiff(x, idxs) | 1.383652 | 1.348899 | 1.364672 | 1.337857 | 1.359290 | 1.334076 |
| 3 | varDiff(x[idxs]) | 1.344870 | 1.347267 | 1.367225 | 1.338651 | 1.373141 | 1.312231 |
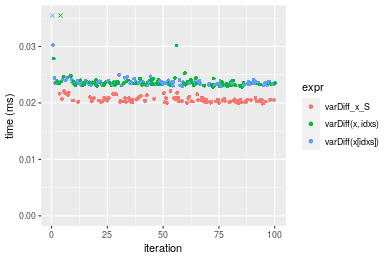
Figure: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on double+n = 10000 data. Outliers are displayed as crosses. Times are in milliseconds.
n = 100000 vector
All elements
> x <- data[["n = 100000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5360272 286.3 7916910 422.9 7916910 422.9
Vcells 11554724 88.2 39038428 297.9 94934136 724.3
> stats <- microbenchmark(varDiff_x_S = varDiff(x_S), `varDiff(x, idxs)` = varDiff(x, idxs = idxs),
+ `varDiff(x[idxs])` = varDiff(x[idxs]), unit = "ms")
Table: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on double+n = 100000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 0.288761 | 0.3207845 | 0.4215957 | 0.3459820 | 0.3860495 | 6.478759 |
| 2 | varDiff(x, idxs) | 0.462206 | 0.4887105 | 0.5596582 | 0.5506450 | 0.5942060 | 0.767178 |
| 3 | varDiff(x[idxs]) | 0.462960 | 0.5058240 | 0.5859137 | 0.5544025 | 0.6596335 | 0.924736 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 2 | varDiff(x, idxs) | 1.600652 | 1.523485 | 1.327476 | 1.591542 | 1.539196 | 0.1184143 |
| 3 | varDiff(x[idxs]) | 1.603264 | 1.576834 | 1.389752 | 1.602403 | 1.708676 | 0.1427335 |
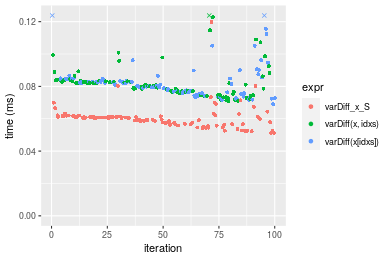
Figure: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on double+n = 100000 data. Outliers are displayed as crosses. Times are in milliseconds.
n = 1000000 vector
All elements
> x <- data[["n = 1000000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5360344 286.3 7916910 422.9 7916910 422.9
Vcells 12500132 95.4 39038428 297.9 94934136 724.3
> stats <- microbenchmark(varDiff_x_S = varDiff(x_S), `varDiff(x, idxs)` = varDiff(x, idxs = idxs),
+ `varDiff(x[idxs])` = varDiff(x[idxs]), unit = "ms")
Table: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on double+n = 1000000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 3.927409 | 4.711675 | 5.06201 | 4.806298 | 4.896624 | 13.03601 |
| 3 | varDiff(x[idxs]) | 10.176256 | 11.106651 | 12.07334 | 11.327246 | 11.979828 | 20.81063 |
| 2 | varDiff(x, idxs) | 10.679407 | 11.178492 | 12.12324 | 11.399456 | 11.807109 | 23.43775 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | varDiff_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 3 | varDiff(x[idxs]) | 2.591086 | 2.357262 | 2.385088 | 2.356750 | 2.446548 | 1.596395 |
| 2 | varDiff(x, idxs) | 2.719199 | 2.372509 | 2.394945 | 2.371774 | 2.411275 | 1.797923 |
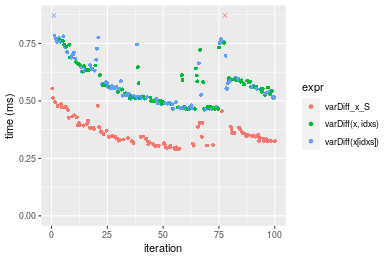
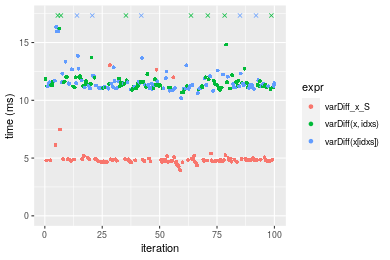
Figure: Benchmarking of varDiff_x_S(), varDiff(x, idxs)() and varDiff(x[idxs])() on double+n = 1000000 data. Outliers are displayed as crosses. Times are in milliseconds.
Appendix
Session information
R version 4.1.1 Patched (2021-08-10 r80727)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.5 LTS
Matrix products: default
BLAS: /home/hb/software/R-devel/R-4-1-branch/lib/R/lib/libRblas.so
LAPACK: /home/hb/software/R-devel/R-4-1-branch/lib/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] microbenchmark_1.4-7 matrixStats_0.60.0 ggplot2_3.3.5
[4] knitr_1.33 R.devices_2.17.0 R.utils_2.10.1
[7] R.oo_1.24.0 R.methodsS3_1.8.1-9001 history_0.0.1-9000
loaded via a namespace (and not attached):
[1] Biobase_2.52.0 httr_1.4.2 splines_4.1.1
[4] bit64_4.0.5 network_1.17.1 assertthat_0.2.1
[7] highr_0.9 stats4_4.1.1 blob_1.2.2
[10] GenomeInfoDbData_1.2.6 robustbase_0.93-8 pillar_1.6.2
[13] RSQLite_2.2.8 lattice_0.20-44 glue_1.4.2
[16] digest_0.6.27 XVector_0.32.0 colorspace_2.0-2
[19] Matrix_1.3-4 XML_3.99-0.7 pkgconfig_2.0.3
[22] zlibbioc_1.38.0 genefilter_1.74.0 purrr_0.3.4
[25] ergm_4.1.2 xtable_1.8-4 scales_1.1.1
[28] tibble_3.1.4 annotate_1.70.0 KEGGREST_1.32.0
[31] farver_2.1.0 generics_0.1.0 IRanges_2.26.0
[34] ellipsis_0.3.2 cachem_1.0.6 withr_2.4.2
[37] BiocGenerics_0.38.0 mime_0.11 survival_3.2-13
[40] magrittr_2.0.1 crayon_1.4.1 statnet.common_4.5.0
[43] memoise_2.0.0 laeken_0.5.1 fansi_0.5.0
[46] R.cache_0.15.0 MASS_7.3-54 R.rsp_0.44.0
[49] progressr_0.8.0 tools_4.1.1 lifecycle_1.0.0
[52] S4Vectors_0.30.0 trust_0.1-8 munsell_0.5.0
[55] tabby_0.0.1-9001 AnnotationDbi_1.54.1 Biostrings_2.60.2
[58] compiler_4.1.1 GenomeInfoDb_1.28.1 rlang_0.4.11
[61] grid_4.1.1 RCurl_1.98-1.4 cwhmisc_6.6
[64] rstudioapi_0.13 rappdirs_0.3.3 startup_0.15.0-9000
[67] labeling_0.4.2 bitops_1.0-7 base64enc_0.1-3
[70] boot_1.3-28 gtable_0.3.0 DBI_1.1.1
[73] markdown_1.1 R6_2.5.1 lpSolveAPI_5.5.2.0-17.7
[76] rle_0.9.2 dplyr_1.0.7 fastmap_1.1.0
[79] bit_4.0.4 utf8_1.2.2 parallel_4.1.1
[82] Rcpp_1.0.7 vctrs_0.3.8 png_0.1-7
[85] DEoptimR_1.0-9 tidyselect_1.1.1 xfun_0.25
[88] coda_0.19-4
Total processing time was 14.48 secs.
Reproducibility
To reproduce this report, do:
html <- matrixStats:::benchmark('varDiff_subset')
Copyright Dongcan Jiang. Last updated on 2021-08-25 22:51:52 (+0200 UTC). Powered by RSP.