matrixStats.benchmarks
colMeans2() and rowMeans2() benchmarks
This report benchmark the performance of colMeans2() and rowMeans2() against alternative methods.
Alternative methods
- apply() + mean()
- .colMeans() and .rowMeans()
- colMeans() and rowMeans()
Data type “integer”
Data
> rmatrix <- function(nrow, ncol, mode = c("logical", "double", "integer", "index"), range = c(-100,
+ +100), na_prob = 0) {
+ mode <- match.arg(mode)
+ n <- nrow * ncol
+ if (mode == "logical") {
+ x <- sample(c(FALSE, TRUE), size = n, replace = TRUE)
+ } else if (mode == "index") {
+ x <- seq_len(n)
+ mode <- "integer"
+ } else {
+ x <- runif(n, min = range[1], max = range[2])
+ }
+ storage.mode(x) <- mode
+ if (na_prob > 0)
+ x[sample(n, size = na_prob * n)] <- NA
+ dim(x) <- c(nrow, ncol)
+ x
+ }
> rmatrices <- function(scale = 10, seed = 1, ...) {
+ set.seed(seed)
+ data <- list()
+ data[[1]] <- rmatrix(nrow = scale * 1, ncol = scale * 1, ...)
+ data[[2]] <- rmatrix(nrow = scale * 10, ncol = scale * 10, ...)
+ data[[3]] <- rmatrix(nrow = scale * 100, ncol = scale * 1, ...)
+ data[[4]] <- t(data[[3]])
+ data[[5]] <- rmatrix(nrow = scale * 10, ncol = scale * 100, ...)
+ data[[6]] <- t(data[[5]])
+ names(data) <- sapply(data, FUN = function(x) paste(dim(x), collapse = "x"))
+ data
+ }
> data <- rmatrices(mode = mode)
Results
10x10 integer matrix
> X <- data[["10x10"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5257921 280.9 7916910 422.9 7916910 422.9
Vcells 10186358 77.8 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5244238 280.1 7916910 422.9 7916910 422.9
Vcells 10141213 77.4 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+10x10 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.002973 | 0.0034290 | 0.0045484 | 0.0039170 | 0.0047025 | 0.029963 |
| 1 | colMeans2 | 0.002586 | 0.0032390 | 0.0050546 | 0.0041735 | 0.0057835 | 0.018183 |
| 3 | colMeans | 0.005649 | 0.0064000 | 0.0090514 | 0.0076845 | 0.0109675 | 0.031805 |
| 4 | apply+mean | 0.070580 | 0.0741775 | 0.0921426 | 0.0758115 | 0.1013795 | 0.227490 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.0000000 | 1.0000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 1 | colMeans2 | 0.8698285 | 0.9445903 | 1.111296 | 1.065484 | 1.229878 | 0.6068484 |
| 3 | colMeans | 1.9001009 | 1.8664334 | 1.990018 | 1.961833 | 2.332270 | 1.0614758 |
| 4 | apply+mean | 23.7403296 | 21.6324001 | 20.258283 | 19.354481 | 21.558639 | 7.5923639 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on integer+10x10 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 0.002397 | 0.0028305 | 0.0035766 | 0.0035125 | 0.0040885 | 0.013986 |
| 2 | .rowMeans | 0.004117 | 0.0045145 | 0.0048169 | 0.0047230 | 0.0049440 | 0.012413 |
| 3 | rowMeans | 0.006448 | 0.0070385 | 0.0080657 | 0.0075265 | 0.0080655 | 0.037236 |
| 4 | apply+mean | 0.067079 | 0.0704940 | 0.0738455 | 0.0736710 | 0.0749055 | 0.139345 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 2 | .rowMeans | 1.717564 | 1.594948 | 1.346761 | 1.344626 | 1.209245 | 0.8875304 |
| 3 | rowMeans | 2.690029 | 2.486663 | 2.255097 | 2.142776 | 1.972728 | 2.6623767 |
| 4 | apply+mean | 27.984564 | 24.905140 | 20.646601 | 20.973950 | 18.321022 | 9.9631775 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+10x10 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
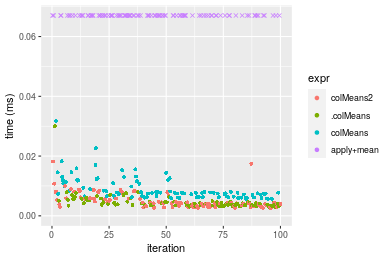
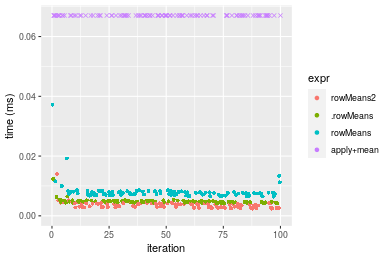 Table: Benchmarking of colMeans2() and rowMeans2() on integer+10x10 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on integer+10x10 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 2.397 | 2.8305 | 3.57664 | 3.5125 | 4.0885 | 13.986 |
| 1 | colMeans2 | 2.586 | 3.2390 | 5.05461 | 4.1735 | 5.7835 | 18.183 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 1 | colMeans2 | 1.078849 | 1.144321 | 1.413229 | 1.188185 | 1.414578 | 1.300086 |
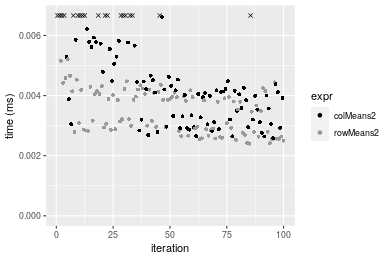
Figure: Benchmarking of colMeans2() and rowMeans2() on integer+10x10 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
100x100 integer matrix
> X <- data[["100x100"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5242821 280.0 7916910 422.9 7916910 422.9
Vcells 9758020 74.5 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5242815 280.0 7916910 422.9 7916910 422.9
Vcells 9763063 74.5 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+100x100 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.012112 | 0.0137450 | 0.0153540 | 0.0146185 | 0.0167420 | 0.024553 |
| 3 | colMeans | 0.014325 | 0.0166630 | 0.0186439 | 0.0177450 | 0.0200885 | 0.041164 |
| 1 | colMeans2 | 0.017660 | 0.0194840 | 0.0213776 | 0.0202760 | 0.0221845 | 0.039210 |
| 4 | apply+mean | 0.425152 | 0.4657975 | 0.5208733 | 0.4980050 | 0.5700890 | 0.776684 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 3 | colMeans | 1.182711 | 1.212295 | 1.214276 | 1.213873 | 1.199887 | 1.676537 |
| 1 | colMeans2 | 1.458058 | 1.417534 | 1.392317 | 1.387010 | 1.325081 | 1.596953 |
| 4 | apply+mean | 35.101717 | 33.888505 | 33.924361 | 34.066765 | 34.051428 | 31.632957 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on integer+100x100 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 0.016040 | 0.0187000 | 0.0217922 | 0.0211005 | 0.0254330 | 0.037635 |
| 2 | .rowMeans | 0.034859 | 0.0369595 | 0.0433372 | 0.0415920 | 0.0463565 | 0.069092 |
| 3 | rowMeans | 0.036387 | 0.0418075 | 0.0465615 | 0.0447965 | 0.0486725 | 0.070743 |
| 4 | apply+mean | 0.400687 | 0.4319210 | 0.4969557 | 0.4827705 | 0.5341675 | 0.750953 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | .rowMeans | 2.173254 | 1.976444 | 1.988656 | 1.971138 | 1.822691 | 1.835844 |
| 3 | rowMeans | 2.268516 | 2.235695 | 2.136609 | 2.123007 | 1.913754 | 1.879713 |
| 4 | apply+mean | 24.980486 | 23.097380 | 22.804260 | 22.879576 | 21.002929 | 19.953580 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+100x100 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
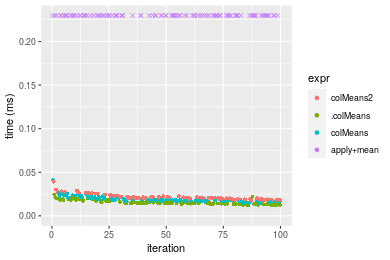
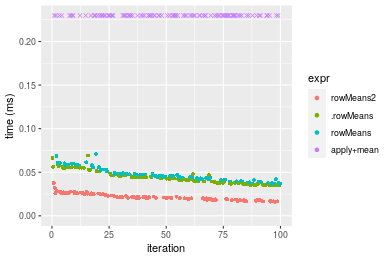 Table: Benchmarking of colMeans2() and rowMeans2() on integer+100x100 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on integer+100x100 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 17.66 | 19.484 | 21.37758 | 20.2760 | 22.1845 | 39.210 |
| 2 | rowMeans2 | 16.04 | 18.700 | 21.79223 | 21.1005 | 25.4330 | 37.635 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 1.0000000 | 1.0000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 2 | rowMeans2 | 0.9082673 | 0.9597619 | 1.019397 | 1.040664 | 1.146431 | 0.9598317 |
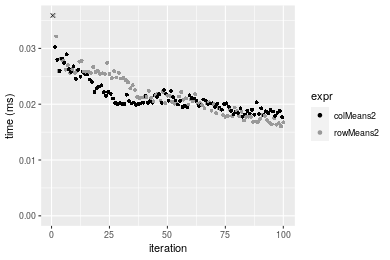
Figure: Benchmarking of colMeans2() and rowMeans2() on integer+100x100 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
1000x10 integer matrix
> X <- data[["1000x10"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5243591 280.1 7916910 422.9 7916910 422.9
Vcells 9761813 74.5 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5243579 280.1 7916910 422.9 7916910 422.9
Vcells 9766846 74.6 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+1000x10 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.013293 | 0.0148880 | 0.0159000 | 0.0159465 | 0.0166735 | 0.022332 |
| 3 | colMeans | 0.016158 | 0.0181375 | 0.0198784 | 0.0192400 | 0.0205770 | 0.048743 |
| 1 | colMeans2 | 0.021063 | 0.0237615 | 0.0250283 | 0.0249125 | 0.0260580 | 0.039862 |
| 4 | apply+mean | 0.140458 | 0.1559735 | 0.1652961 | 0.1653795 | 0.1731740 | 0.278474 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 3 | colMeans | 1.215527 | 1.218263 | 1.250215 | 1.206534 | 1.234114 | 2.182653 |
| 1 | colMeans2 | 1.584518 | 1.596017 | 1.574109 | 1.562255 | 1.562839 | 1.784972 |
| 4 | apply+mean | 10.566313 | 10.476458 | 10.395984 | 10.370896 | 10.386182 | 12.469729 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on integer+1000x10 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 0.018759 | 0.0205295 | 0.0230194 | 0.0216080 | 0.0234490 | 0.043084 |
| 4 | apply+mean | 0.124769 | 0.1375675 | 0.1546701 | 0.1434855 | 0.1663905 | 0.291129 |
| 2 | .rowMeans | 0.156096 | 0.1666985 | 0.1835071 | 0.1722290 | 0.2058055 | 0.232899 |
| 3 | rowMeans | 0.157676 | 0.1692335 | 0.1852035 | 0.1749680 | 0.1966100 | 0.259823 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 4 | apply+mean | 6.651154 | 6.700967 | 6.719132 | 6.640388 | 7.095846 | 6.757242 |
| 2 | .rowMeans | 8.321126 | 8.119949 | 7.971860 | 7.970613 | 8.776728 | 5.405696 |
| 3 | rowMeans | 8.405352 | 8.243430 | 8.045555 | 8.097371 | 8.384579 | 6.030615 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+1000x10 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
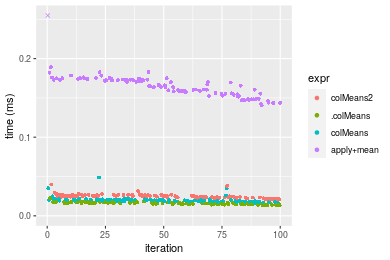
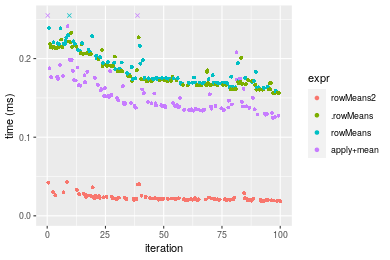 Table: Benchmarking of colMeans2() and rowMeans2() on integer+1000x10 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on integer+1000x10 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 18.759 | 20.5295 | 23.01936 | 21.6080 | 23.449 | 43.084 |
| 1 | colMeans2 | 21.063 | 23.7615 | 25.02834 | 24.9125 | 26.058 | 39.862 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 1 | colMeans2 | 1.122821 | 1.157432 | 1.087274 | 1.152929 | 1.111263 | 0.9252159 |
Figure: Benchmarking of colMeans2() and rowMeans2() on integer+1000x10 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
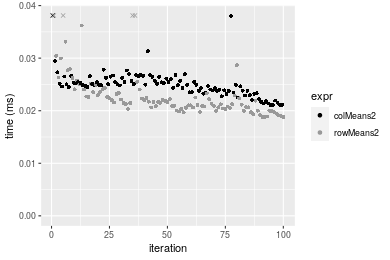
10x1000 integer matrix
> X <- data[["10x1000"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5243807 280.1 7916910 422.9 7916910 422.9
Vcells 9762612 74.5 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5243801 280.1 7916910 422.9 7916910 422.9
Vcells 9767655 74.6 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+10x1000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.012298 | 0.0140630 | 0.0160430 | 0.0157990 | 0.0168240 | 0.026693 |
| 3 | colMeans | 0.014148 | 0.0169780 | 0.0213489 | 0.0190300 | 0.0230850 | 0.068485 |
| 1 | colMeans2 | 0.015985 | 0.0180765 | 0.0211984 | 0.0196635 | 0.0227085 | 0.044537 |
| 4 | apply+mean | 3.091406 | 3.2096035 | 3.6078381 | 3.4905205 | 3.7838090 | 8.776626 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 3 | colMeans | 1.150431 | 1.207281 | 1.330730 | 1.204507 | 1.372147 | 2.565654 |
| 1 | colMeans2 | 1.299805 | 1.285394 | 1.321347 | 1.244604 | 1.349768 | 1.668490 |
| 4 | apply+mean | 251.374695 | 228.230356 | 224.884938 | 220.933002 | 224.905433 | 328.798786 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on integer+10x1000 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 0.015877 | 0.0166955 | 0.0218266 | 0.019220 | 0.0250790 | 0.064051 |
| 2 | .rowMeans | 0.026027 | 0.0264320 | 0.0293154 | 0.028028 | 0.0307880 | 0.051111 |
| 3 | rowMeans | 0.027577 | 0.0289865 | 0.0340805 | 0.031922 | 0.0356905 | 0.064902 |
| 4 | apply+mean | 3.082995 | 3.2155610 | 3.5407100 | 3.380379 | 3.5959655 | 8.124443 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 2 | .rowMeans | 1.639290 | 1.583181 | 1.343103 | 1.458273 | 1.227641 | 0.7979735 |
| 3 | rowMeans | 1.736915 | 1.736186 | 1.561421 | 1.660874 | 1.423123 | 1.0132863 |
| 4 | apply+mean | 194.179946 | 192.600461 | 162.219804 | 175.878200 | 143.385522 | 126.8433436 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+10x1000 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
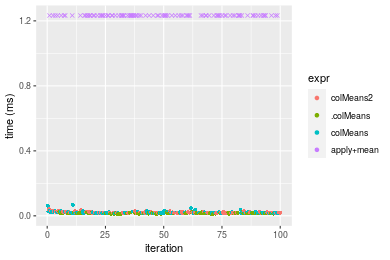
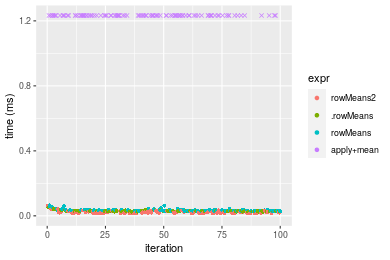 Table: Benchmarking of colMeans2() and rowMeans2() on integer+10x1000 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on integer+10x1000 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 15.877 | 16.6955 | 21.82662 | 19.2200 | 25.0790 | 64.051 |
| 1 | colMeans2 | 15.985 | 18.0765 | 21.19842 | 19.6635 | 22.7085 | 44.537 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 1.000000 | 1.000000 | 1.0000000 | 1.000000 | 1.0000000 | 1.0000000 |
| 1 | colMeans2 | 1.006802 | 1.082717 | 0.9712186 | 1.023075 | 0.9054787 | 0.6953365 |
Figure: Benchmarking of colMeans2() and rowMeans2() on integer+10x1000 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
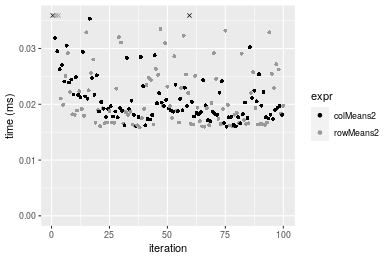
100x1000 integer matrix
> X <- data[["100x1000"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5244037 280.1 7916910 422.9 7916910 422.9
Vcells 9763204 74.5 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5244031 280.1 7916910 422.9 7916910 422.9
Vcells 9813247 74.9 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+100x1000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.088433 | 0.0888500 | 0.0948695 | 0.0903635 | 0.0930635 | 0.171991 |
| 3 | colMeans | 0.090535 | 0.0911145 | 0.0972239 | 0.0930615 | 0.0978145 | 0.142057 |
| 1 | colMeans2 | 0.138827 | 0.1393920 | 0.1472027 | 0.1408785 | 0.1454935 | 0.211462 |
| 4 | apply+mean | 3.670839 | 3.7336380 | 4.1002198 | 3.8298305 | 3.9675910 | 16.060589 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 3 | colMeans | 1.023769 | 1.025487 | 1.024817 | 1.029857 | 1.051051 | 0.825956 |
| 1 | colMeans2 | 1.569855 | 1.568846 | 1.551633 | 1.559020 | 1.563379 | 1.229495 |
| 4 | apply+mean | 41.509832 | 42.021812 | 43.219574 | 42.382494 | 42.633159 | 93.380404 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on integer+100x1000 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 0.139095 | 0.1402800 | 0.1533820 | 0.1429255 | 0.1535730 | 0.238553 |
| 2 | .rowMeans | 0.221425 | 0.2220600 | 0.2383832 | 0.2241970 | 0.2349075 | 0.381985 |
| 3 | rowMeans | 0.223737 | 0.2252015 | 0.2397627 | 0.2293305 | 0.2426695 | 0.311189 |
| 4 | apply+mean | 3.674989 | 3.7546890 | 4.1612656 | 3.8256255 | 4.0240735 | 19.497721 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | .rowMeans | 1.591898 | 1.582977 | 1.554180 | 1.568628 | 1.529615 | 1.601258 |
| 3 | rowMeans | 1.608519 | 1.605371 | 1.563173 | 1.604546 | 1.580157 | 1.304486 |
| 4 | apply+mean | 26.420713 | 26.765676 | 27.130078 | 26.766571 | 26.203001 | 81.733288 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+100x1000 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
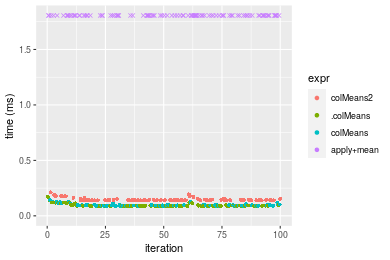
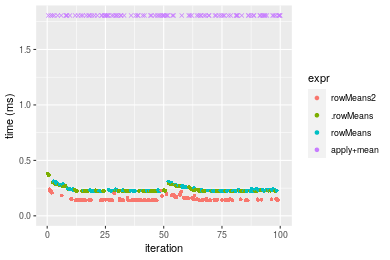 Table: Benchmarking of colMeans2() and rowMeans2() on integer+100x1000 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on integer+100x1000 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 138.827 | 139.392 | 147.2027 | 140.8785 | 145.4935 | 211.462 |
| 2 | rowMeans2 | 139.095 | 140.280 | 153.3820 | 142.9255 | 153.5730 | 238.553 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.00000 | 1.000000 | 1.000000 |
| 2 | rowMeans2 | 1.001931 | 1.006371 | 1.041978 | 1.01453 | 1.055532 | 1.128113 |
Figure: Benchmarking of colMeans2() and rowMeans2() on integer+100x1000 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
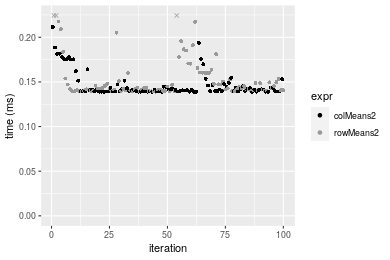
1000x100 integer matrix
> X <- data[["1000x100"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5244261 280.1 7916910 422.9 7916910 422.9
Vcells 9763984 74.5 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5244255 280.1 7916910 422.9 7916910 422.9
Vcells 9814027 74.9 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+1000x100 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.081000 | 0.0813370 | 0.0899024 | 0.0821110 | 0.0887765 | 0.148613 |
| 3 | colMeans | 0.083234 | 0.0838735 | 0.0939699 | 0.0853420 | 0.1025235 | 0.146897 |
| 1 | colMeans2 | 0.132713 | 0.1333555 | 0.1479354 | 0.1344490 | 0.1597905 | 0.230693 |
| 4 | apply+mean | 0.893604 | 0.9013625 | 1.0113902 | 0.9101045 | 1.0796735 | 1.572386 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 3 | colMeans | 1.027580 | 1.031185 | 1.045244 | 1.039349 | 1.154850 | 0.9884532 |
| 1 | colMeans2 | 1.638432 | 1.639543 | 1.645511 | 1.637406 | 1.799919 | 1.5523070 |
| 4 | apply+mean | 11.032148 | 11.081826 | 11.249871 | 11.083832 | 12.161704 | 10.5804068 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on integer+1000x100 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 0.134541 | 0.1355810 | 0.1441573 | 0.1367815 | 0.139278 | 0.225825 |
| 2 | .rowMeans | 0.320036 | 0.3209695 | 0.3420746 | 0.3212865 | 0.327356 | 0.541075 |
| 3 | rowMeans | 0.322424 | 0.3235910 | 0.3447219 | 0.3245065 | 0.345412 | 0.514245 |
| 4 | apply+mean | 0.893850 | 0.9032450 | 0.9761286 | 0.9099045 | 0.950947 | 1.584763 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | .rowMeans | 2.378725 | 2.367363 | 2.372926 | 2.348903 | 2.350378 | 2.395993 |
| 3 | rowMeans | 2.396474 | 2.386699 | 2.391290 | 2.372444 | 2.480018 | 2.277184 |
| 4 | apply+mean | 6.643700 | 6.662032 | 6.771274 | 6.652248 | 6.827690 | 7.017660 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on integer+1000x100 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
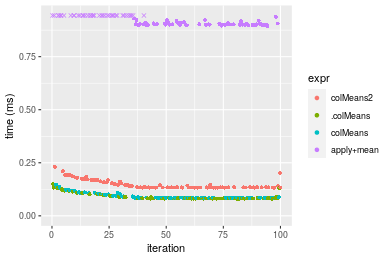
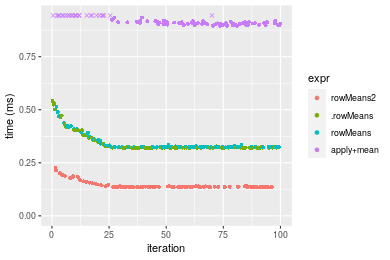 Table: Benchmarking of colMeans2() and rowMeans2() on integer+1000x100 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on integer+1000x100 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 132.713 | 133.3555 | 147.9354 | 134.4490 | 159.7905 | 230.693 |
| 2 | rowMeans2 | 134.541 | 135.5810 | 144.1573 | 136.7815 | 139.2780 | 225.825 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 1.000000 | 1.000000 | 1.0000000 | 1.000000 | 1.0000000 | 1.0000000 |
| 2 | rowMeans2 | 1.013774 | 1.016689 | 0.9744613 | 1.017349 | 0.8716288 | 0.9788984 |
Figure: Benchmarking of colMeans2() and rowMeans2() on integer+1000x100 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
Data type “double”
Data
> rmatrix <- function(nrow, ncol, mode = c("logical", "double", "integer", "index"), range = c(-100,
+ +100), na_prob = 0) {
+ mode <- match.arg(mode)
+ n <- nrow * ncol
+ if (mode == "logical") {
+ x <- sample(c(FALSE, TRUE), size = n, replace = TRUE)
+ } else if (mode == "index") {
+ x <- seq_len(n)
+ mode <- "integer"
+ } else {
+ x <- runif(n, min = range[1], max = range[2])
+ }
+ storage.mode(x) <- mode
+ if (na_prob > 0)
+ x[sample(n, size = na_prob * n)] <- NA
+ dim(x) <- c(nrow, ncol)
+ x
+ }
> rmatrices <- function(scale = 10, seed = 1, ...) {
+ set.seed(seed)
+ data <- list()
+ data[[1]] <- rmatrix(nrow = scale * 1, ncol = scale * 1, ...)
+ data[[2]] <- rmatrix(nrow = scale * 10, ncol = scale * 10, ...)
+ data[[3]] <- rmatrix(nrow = scale * 100, ncol = scale * 1, ...)
+ data[[4]] <- t(data[[3]])
+ data[[5]] <- rmatrix(nrow = scale * 10, ncol = scale * 100, ...)
+ data[[6]] <- t(data[[5]])
+ names(data) <- sapply(data, FUN = function(x) paste(dim(x), collapse = "x"))
+ data
+ }
> data <- rmatrices(mode = mode)
Results
10x10 double matrix
> X <- data[["10x10"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5244498 280.1 7916910 422.9 7916910 422.9
Vcells 9879920 75.4 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5244477 280.1 7916910 422.9 7916910 422.9
Vcells 9880038 75.4 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+10x10 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.002776 | 0.0031885 | 0.0037724 | 0.003554 | 0.0041845 | 0.013491 |
| 1 | colMeans2 | 0.002411 | 0.0027805 | 0.0036513 | 0.003734 | 0.0041320 | 0.015417 |
| 3 | colMeans | 0.005321 | 0.0058290 | 0.0067177 | 0.006375 | 0.0074175 | 0.018090 |
| 4 | apply+mean | 0.068469 | 0.0712470 | 0.0744565 | 0.073194 | 0.0750125 | 0.155593 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.0000000 | 1.0000000 | 1.0000000 | 1.000000 | 1.0000000 | 1.000000 |
| 1 | colMeans2 | 0.8685159 | 0.8720401 | 0.9679089 | 1.050647 | 0.9874537 | 1.142762 |
| 3 | colMeans | 1.9167867 | 1.8281324 | 1.7807697 | 1.793754 | 1.7726132 | 1.340894 |
| 4 | apply+mean | 24.6646254 | 22.3449898 | 19.7372720 | 20.594823 | 17.9262755 | 11.533096 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on double+10x10 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .rowMeans | 0.002426 | 0.0027290 | 0.0030330 | 0.0029570 | 0.0031865 | 0.007529 |
| 1 | rowMeans2 | 0.002355 | 0.0028005 | 0.0035996 | 0.0036665 | 0.0041455 | 0.014751 |
| 3 | rowMeans | 0.004684 | 0.0051880 | 0.0060911 | 0.0059685 | 0.0063850 | 0.027172 |
| 4 | apply+mean | 0.066863 | 0.0715830 | 0.0743118 | 0.0737275 | 0.0747935 | 0.135378 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .rowMeans | 1.0000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 1 | rowMeans2 | 0.9707337 | 1.026200 | 1.186844 | 1.239939 | 1.300957 | 1.959224 |
| 3 | rowMeans | 1.9307502 | 1.901063 | 2.008305 | 2.018431 | 2.003766 | 3.608979 |
| 4 | apply+mean | 27.5610058 | 26.230487 | 24.501395 | 24.933209 | 23.471991 | 17.980874 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+10x10 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
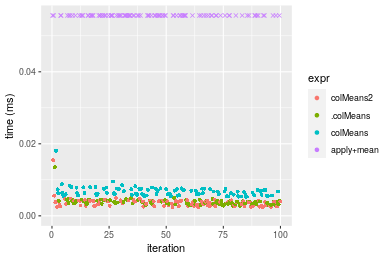
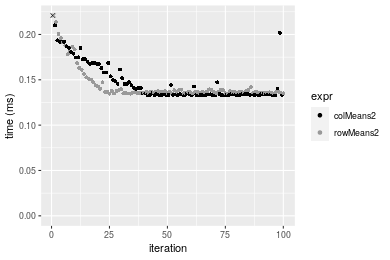
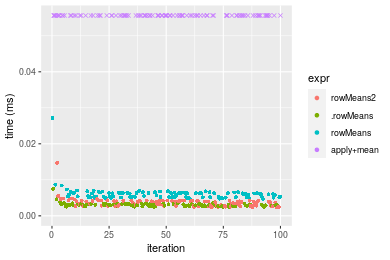 Table: Benchmarking of colMeans2() and rowMeans2() on double+10x10 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on double+10x10 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 2.355 | 2.8005 | 3.59965 | 3.6665 | 4.1455 | 14.751 |
| 1 | colMeans2 | 2.411 | 2.7805 | 3.65132 | 3.7340 | 4.1320 | 15.417 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 1.000000 | 1.0000000 | 1.000000 | 1.00000 | 1.0000000 | 1.000000 |
| 1 | colMeans2 | 1.023779 | 0.9928584 | 1.014354 | 1.01841 | 0.9967435 | 1.045149 |
Figure: Benchmarking of colMeans2() and rowMeans2() on double+10x10 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
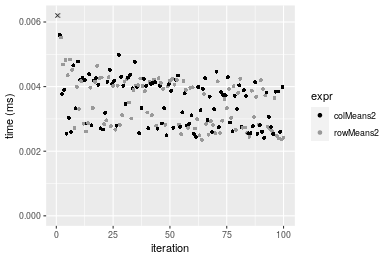
100x100 double matrix
> X <- data[["100x100"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5244711 280.1 7916910 422.9 7916910 422.9
Vcells 9880943 75.4 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5244705 280.1 7916910 422.9 7916910 422.9
Vcells 9890986 75.5 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+100x100 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.009332 | 0.0103345 | 0.0116088 | 0.0112220 | 0.0122990 | 0.019403 |
| 3 | colMeans | 0.011515 | 0.0132410 | 0.0149219 | 0.0140775 | 0.0158050 | 0.039144 |
| 1 | colMeans2 | 0.013225 | 0.0147725 | 0.0165608 | 0.0157235 | 0.0170340 | 0.034387 |
| 4 | apply+mean | 0.425886 | 0.4646460 | 0.5187741 | 0.4955835 | 0.5462565 | 0.796944 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 3 | colMeans | 1.233926 | 1.281242 | 1.285392 | 1.254455 | 1.285064 | 2.017420 |
| 1 | colMeans2 | 1.417167 | 1.429435 | 1.426566 | 1.401132 | 1.384991 | 1.772252 |
| 4 | apply+mean | 45.637163 | 44.960666 | 44.687890 | 44.161780 | 44.414709 | 41.073236 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on double+100x100 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 0.013392 | 0.0159650 | 0.0186733 | 0.0179485 | 0.0203945 | 0.032255 |
| 2 | .rowMeans | 0.025087 | 0.0273720 | 0.0308377 | 0.0297420 | 0.0340090 | 0.049206 |
| 3 | rowMeans | 0.027367 | 0.0300700 | 0.0338425 | 0.0324820 | 0.0369925 | 0.053002 |
| 4 | apply+mean | 0.425429 | 0.4658275 | 0.5324830 | 0.5030520 | 0.5608050 | 0.929950 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | .rowMeans | 1.873283 | 1.714500 | 1.651431 | 1.657074 | 1.667557 | 1.525531 |
| 3 | rowMeans | 2.043534 | 1.883495 | 1.812346 | 1.809733 | 1.813847 | 1.643218 |
| 4 | apply+mean | 31.767398 | 29.178046 | 28.515725 | 28.027523 | 27.497855 | 28.831189 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+100x100 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
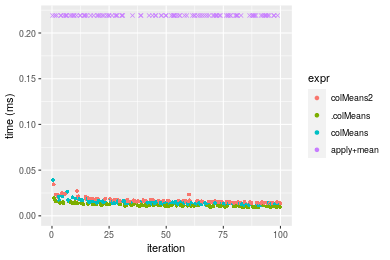
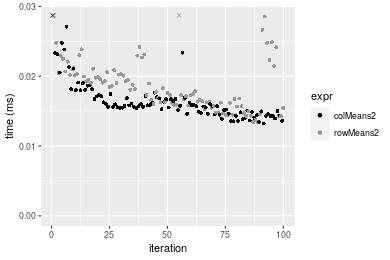
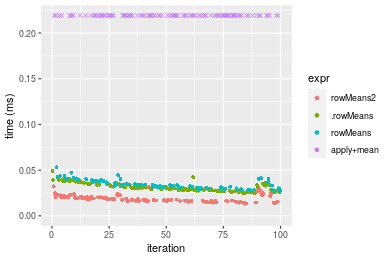 Table: Benchmarking of colMeans2() and rowMeans2() on double+100x100 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on double+100x100 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 13.225 | 14.7725 | 16.56076 | 15.7235 | 17.0340 | 34.387 |
| 2 | rowMeans2 | 13.392 | 15.9650 | 18.67331 | 17.9485 | 20.3945 | 32.255 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 2 | rowMeans2 | 1.012628 | 1.080724 | 1.127564 | 1.141508 | 1.197282 | 0.9379998 |
Figure: Benchmarking of colMeans2() and rowMeans2() on double+100x100 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
1000x10 double matrix
> X <- data[["1000x10"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5244941 280.2 7916910 422.9 7916910 422.9
Vcells 9882068 75.4 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5244929 280.2 7916910 422.9 7916910 422.9
Vcells 9892101 75.5 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+1000x10 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.012477 | 0.0136280 | 0.0150680 | 0.0147515 | 0.0159775 | 0.023577 |
| 3 | colMeans | 0.015002 | 0.0168430 | 0.0191004 | 0.0181305 | 0.0195950 | 0.050398 |
| 1 | colMeans2 | 0.016514 | 0.0184595 | 0.0206924 | 0.0205380 | 0.0215860 | 0.035745 |
| 4 | apply+mean | 0.151964 | 0.1621315 | 0.1802185 | 0.1756700 | 0.1934355 | 0.320478 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 3 | colMeans | 1.202372 | 1.235911 | 1.267612 | 1.229062 | 1.226412 | 2.137592 |
| 1 | colMeans2 | 1.323555 | 1.354527 | 1.373266 | 1.392265 | 1.351025 | 1.516096 |
| 4 | apply+mean | 12.179530 | 11.896940 | 11.960325 | 11.908620 | 12.106744 | 13.592823 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on double+1000x10 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 0.016585 | 0.0176235 | 0.0193872 | 0.0190780 | 0.0201155 | 0.036711 |
| 2 | .rowMeans | 0.029477 | 0.0310105 | 0.0337392 | 0.0329205 | 0.0356865 | 0.046924 |
| 3 | rowMeans | 0.031684 | 0.0339570 | 0.0370014 | 0.0355740 | 0.0390985 | 0.057199 |
| 4 | apply+mean | 0.146000 | 0.1541335 | 0.1709902 | 0.1683030 | 0.1799280 | 0.286069 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | .rowMeans | 1.777329 | 1.759611 | 1.740279 | 1.725574 | 1.774080 | 1.278200 |
| 3 | rowMeans | 1.910401 | 1.926802 | 1.908546 | 1.864661 | 1.943700 | 1.558089 |
| 4 | apply+mean | 8.803135 | 8.745907 | 8.819727 | 8.821837 | 8.944744 | 7.792460 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+1000x10 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
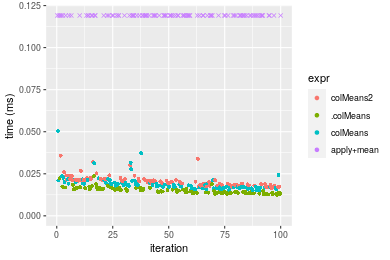
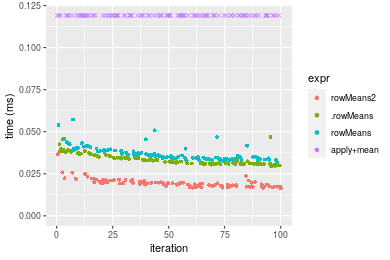 Table: Benchmarking of colMeans2() and rowMeans2() on double+1000x10 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on double+1000x10 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 16.585 | 17.6235 | 19.38724 | 19.078 | 20.1155 | 36.711 |
| 1 | colMeans2 | 16.514 | 18.4595 | 20.69241 | 20.538 | 21.5860 | 35.745 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 1 | colMeans2 | 0.995719 | 1.047437 | 1.067321 | 1.076528 | 1.073103 | 0.9736864 |
Figure: Benchmarking of colMeans2() and rowMeans2() on double+1000x10 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
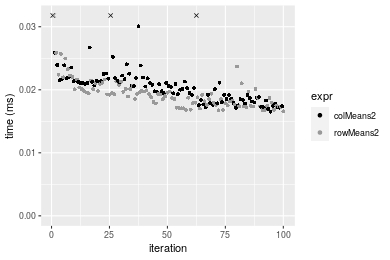
10x1000 double matrix
> X <- data[["10x1000"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5245157 280.2 7916910 422.9 7916910 422.9
Vcells 9882205 75.4 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5245151 280.2 7916910 422.9 7916910 422.9
Vcells 9892248 75.5 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+10x1000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.009626 | 0.0101775 | 0.0120814 | 0.0115835 | 0.0129925 | 0.025867 |
| 3 | colMeans | 0.011458 | 0.0125005 | 0.0162083 | 0.0145280 | 0.0173725 | 0.053365 |
| 1 | colMeans2 | 0.014976 | 0.0158040 | 0.0186646 | 0.0171290 | 0.0194755 | 0.048990 |
| 4 | apply+mean | 3.086088 | 3.1559520 | 3.4518498 | 3.2799350 | 3.4935680 | 8.668491 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 3 | colMeans | 1.190318 | 1.228249 | 1.341593 | 1.254198 | 1.337118 | 2.063053 |
| 1 | colMeans2 | 1.555786 | 1.552837 | 1.544902 | 1.478741 | 1.498980 | 1.893919 |
| 4 | apply+mean | 320.599211 | 310.091083 | 285.715807 | 283.155782 | 268.891129 | 335.117756 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on double+10x1000 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 0.015009 | 0.0156130 | 0.0190411 | 0.0169375 | 0.0195240 | 0.057578 |
| 2 | .rowMeans | 0.025013 | 0.0254050 | 0.0275033 | 0.0263310 | 0.0281965 | 0.048781 |
| 3 | rowMeans | 0.026902 | 0.0278405 | 0.0311949 | 0.0295030 | 0.0322195 | 0.060199 |
| 4 | apply+mean | 3.069896 | 3.1930860 | 3.4740875 | 3.2728830 | 3.4834760 | 8.631549 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | .rowMeans | 1.666533 | 1.627170 | 1.444414 | 1.554598 | 1.444197 | 0.847216 |
| 3 | rowMeans | 1.792391 | 1.783162 | 1.638293 | 1.741874 | 1.650251 | 1.045521 |
| 4 | apply+mean | 204.537011 | 204.514571 | 182.451844 | 193.232945 | 178.420201 | 149.910539 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+10x1000 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
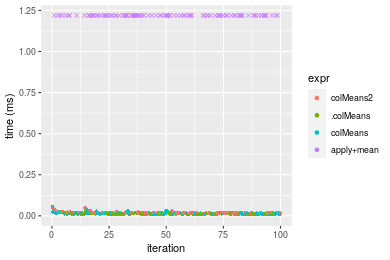
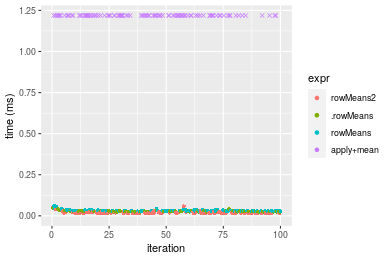 Table: Benchmarking of colMeans2() and rowMeans2() on double+10x1000 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on double+10x1000 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 15.009 | 15.613 | 19.04112 | 16.9375 | 19.5240 | 57.578 |
| 1 | colMeans2 | 14.976 | 15.804 | 18.66459 | 17.1290 | 19.4755 | 48.990 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | rowMeans2 | 1.0000000 | 1.000000 | 1.0000000 | 1.000000 | 1.0000000 | 1.0000000 |
| 1 | colMeans2 | 0.9978013 | 1.012233 | 0.9802254 | 1.011306 | 0.9975159 | 0.8508458 |
Figure: Benchmarking of colMeans2() and rowMeans2() on double+10x1000 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
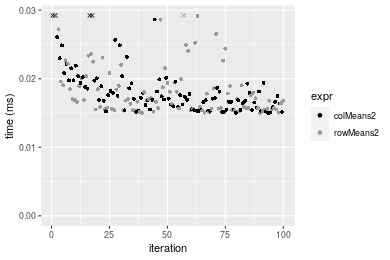
100x1000 double matrix
> X <- data[["100x1000"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5245387 280.2 7916910 422.9 7916910 422.9
Vcells 9883526 75.5 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5245381 280.2 7916910 422.9 7916910 422.9
Vcells 9983569 76.2 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+100x1000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.063167 | 0.0638000 | 0.0737637 | 0.0674975 | 0.0780270 | 0.159803 |
| 3 | colMeans | 0.065253 | 0.0660820 | 0.0755566 | 0.0693515 | 0.0817260 | 0.142163 |
| 1 | colMeans2 | 0.104201 | 0.1049505 | 0.1161812 | 0.1094545 | 0.1198685 | 0.179286 |
| 4 | apply+mean | 3.785904 | 3.8621365 | 4.3237555 | 4.0004650 | 4.1720880 | 18.349369 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 3 | colMeans | 1.033024 | 1.035768 | 1.024306 | 1.027468 | 1.047407 | 0.8896141 |
| 1 | colMeans2 | 1.649611 | 1.644992 | 1.575044 | 1.621608 | 1.536244 | 1.1219189 |
| 4 | apply+mean | 59.934839 | 60.535055 | 58.616272 | 59.268343 | 53.469799 | 114.8249345 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on double+100x1000 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 0.109627 | 0.1129705 | 0.1305515 | 0.1195635 | 0.1331590 | 0.369935 |
| 2 | .rowMeans | 0.210915 | 0.2115055 | 0.2348538 | 0.2157175 | 0.2437235 | 0.409699 |
| 3 | rowMeans | 0.213266 | 0.2142505 | 0.2363136 | 0.2187875 | 0.2484610 | 0.382752 |
| 4 | apply+mean | 3.804420 | 3.8713845 | 4.3908263 | 3.9796310 | 4.1606125 | 20.476403 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | .rowMeans | 1.923933 | 1.872219 | 1.798936 | 1.804209 | 1.830319 | 1.107489 |
| 3 | rowMeans | 1.945378 | 1.896517 | 1.810118 | 1.829885 | 1.865897 | 1.034647 |
| 4 | apply+mean | 34.703312 | 34.268986 | 33.632908 | 33.284665 | 31.245447 | 55.351354 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+100x1000 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
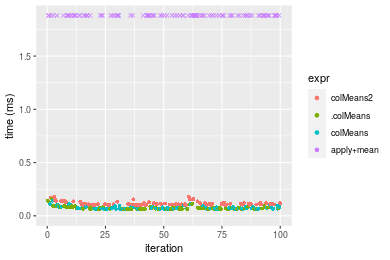
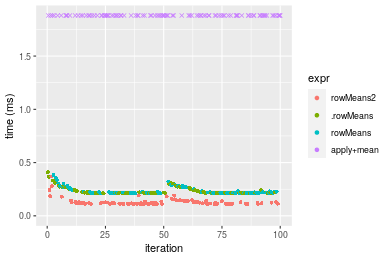 Table: Benchmarking of colMeans2() and rowMeans2() on double+100x1000 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on double+100x1000 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 104.201 | 104.9505 | 116.1812 | 109.4545 | 119.8685 | 179.286 |
| 2 | rowMeans2 | 109.627 | 112.9705 | 130.5515 | 119.5635 | 133.1590 | 369.935 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | rowMeans2 | 1.052072 | 1.076417 | 1.123689 | 1.092358 | 1.110876 | 2.063379 |
Figure: Benchmarking of colMeans2() and rowMeans2() on double+100x1000 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
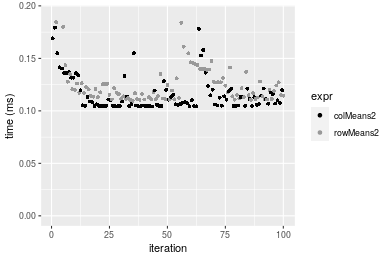
1000x100 double matrix
> X <- data[["1000x100"]]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5245611 280.2 7916910 422.9 7916910 422.9
Vcells 9883675 75.5 33191153 253.3 53339345 407.0
> colStats <- microbenchmark(colMeans2 = colMeans2(X, na.rm = FALSE), .colMeans = .colMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), colMeans = colMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 2L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
> X <- t(X)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5245605 280.2 7916910 422.9 7916910 422.9
Vcells 9983718 76.2 33191153 253.3 53339345 407.0
> rowStats <- microbenchmark(rowMeans2 = rowMeans2(X, na.rm = FALSE), .rowMeans = .rowMeans(X, m = nrow(X),
+ n = ncol(X), na.rm = FALSE), rowMeans = rowMeans(X, na.rm = FALSE), `apply+mean` = apply(X, MARGIN = 1L,
+ FUN = mean, na.rm = FALSE), unit = "ms")
Table: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+1000x100 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 0.078013 | 0.078512 | 0.0923077 | 0.080634 | 0.1019995 | 0.159646 |
| 3 | colMeans | 0.080225 | 0.081468 | 0.0958915 | 0.089389 | 0.1062610 | 0.151461 |
| 1 | colMeans2 | 0.106241 | 0.108208 | 0.1251798 | 0.118682 | 0.1379010 | 0.185638 |
| 4 | apply+mean | 0.999177 | 1.033953 | 1.2982486 | 1.196687 | 1.3312755 | 8.843105 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 2 | .colMeans | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.0000000 |
| 3 | colMeans | 1.028354 | 1.037650 | 1.038824 | 1.108577 | 1.041780 | 0.9487303 |
| 1 | colMeans2 | 1.361837 | 1.378235 | 1.356114 | 1.471861 | 1.351977 | 1.1628102 |
| 4 | apply+mean | 12.807827 | 13.169356 | 14.064354 | 14.840979 | 13.051785 | 55.3919610 |
Table: Benchmarking of rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on double+1000x100 data (transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 0.110857 | 0.1143645 | 0.1347093 | 0.1257395 | 0.1430745 | 0.249168 |
| 2 | .rowMeans | 0.214976 | 0.2158880 | 0.2522388 | 0.2332150 | 0.2794755 | 0.396863 |
| 3 | rowMeans | 0.217536 | 0.2199010 | 0.2625061 | 0.2465075 | 0.2836185 | 0.410389 |
| 4 | apply+mean | 1.028700 | 1.0431015 | 1.3299909 | 1.1217340 | 1.3888410 | 8.272883 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | rowMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | .rowMeans | 1.939219 | 1.887719 | 1.872468 | 1.854747 | 1.953356 | 1.592753 |
| 3 | rowMeans | 1.962312 | 1.922808 | 1.948686 | 1.960462 | 1.982313 | 1.647037 |
| 4 | apply+mean | 9.279522 | 9.120850 | 9.873043 | 8.921095 | 9.707118 | 33.202028 |
Figure: Benchmarking of colMeans2(), .colMeans(), colMeans() and apply+mean() on double+1000x100 data as well as rowMeans2(), .rowMeans(), rowMeans() and apply+mean() on the same data transposed. Outliers are displayed as crosses. Times are in milliseconds.
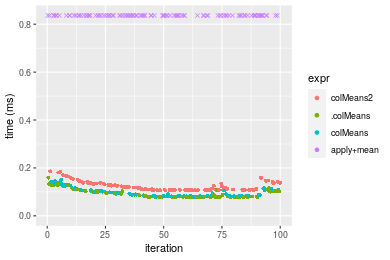
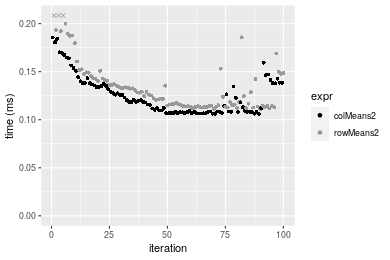
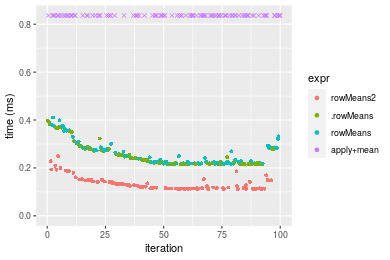 Table: Benchmarking of colMeans2() and rowMeans2() on double+1000x100 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
Table: Benchmarking of colMeans2() and rowMeans2() on double+1000x100 data (original and transposed). The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 106.241 | 108.2080 | 125.1798 | 118.6820 | 137.9010 | 185.638 |
| 2 | rowMeans2 | 110.857 | 114.3645 | 134.7093 | 125.7395 | 143.0745 | 249.168 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | colMeans2 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | rowMeans2 | 1.043448 | 1.056895 | 1.076126 | 1.059466 | 1.037516 | 1.342225 |
Figure: Benchmarking of colMeans2() and rowMeans2() on double+1000x100 data (original and transposed). Outliers are displayed as crosses. Times are in milliseconds.
Appendix
Session information
R version 4.1.1 Patched (2021-08-10 r80727)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.5 LTS
Matrix products: default
BLAS: /home/hb/software/R-devel/R-4-1-branch/lib/R/lib/libRblas.so
LAPACK: /home/hb/software/R-devel/R-4-1-branch/lib/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] microbenchmark_1.4-7 matrixStats_0.60.0 ggplot2_3.3.5
[4] knitr_1.33 R.devices_2.17.0 R.utils_2.10.1
[7] R.oo_1.24.0 R.methodsS3_1.8.1-9001 history_0.0.1-9000
loaded via a namespace (and not attached):
[1] Biobase_2.52.0 httr_1.4.2 splines_4.1.1
[4] bit64_4.0.5 network_1.17.1 assertthat_0.2.1
[7] highr_0.9 stats4_4.1.1 blob_1.2.2
[10] GenomeInfoDbData_1.2.6 robustbase_0.93-8 pillar_1.6.2
[13] RSQLite_2.2.8 lattice_0.20-44 glue_1.4.2
[16] digest_0.6.27 XVector_0.32.0 colorspace_2.0-2
[19] Matrix_1.3-4 XML_3.99-0.7 pkgconfig_2.0.3
[22] zlibbioc_1.38.0 genefilter_1.74.0 purrr_0.3.4
[25] ergm_4.1.2 xtable_1.8-4 scales_1.1.1
[28] tibble_3.1.4 annotate_1.70.0 KEGGREST_1.32.0
[31] farver_2.1.0 generics_0.1.0 IRanges_2.26.0
[34] ellipsis_0.3.2 cachem_1.0.6 withr_2.4.2
[37] BiocGenerics_0.38.0 mime_0.11 survival_3.2-13
[40] magrittr_2.0.1 crayon_1.4.1 statnet.common_4.5.0
[43] memoise_2.0.0 laeken_0.5.1 fansi_0.5.0
[46] R.cache_0.15.0 MASS_7.3-54 R.rsp_0.44.0
[49] progressr_0.8.0 tools_4.1.1 lifecycle_1.0.0
[52] S4Vectors_0.30.0 trust_0.1-8 munsell_0.5.0
[55] tabby_0.0.1-9001 AnnotationDbi_1.54.1 Biostrings_2.60.2
[58] compiler_4.1.1 GenomeInfoDb_1.28.1 rlang_0.4.11
[61] grid_4.1.1 RCurl_1.98-1.4 cwhmisc_6.6
[64] rstudioapi_0.13 rappdirs_0.3.3 startup_0.15.0
[67] labeling_0.4.2 bitops_1.0-7 base64enc_0.1-3
[70] boot_1.3-28 gtable_0.3.0 DBI_1.1.1
[73] markdown_1.1 R6_2.5.1 lpSolveAPI_5.5.2.0-17.7
[76] rle_0.9.2 dplyr_1.0.7 fastmap_1.1.0
[79] bit_4.0.4 utf8_1.2.2 parallel_4.1.1
[82] Rcpp_1.0.7 vctrs_0.3.8 png_0.1-7
[85] DEoptimR_1.0-9 tidyselect_1.1.1 xfun_0.25
[88] coda_0.19-4
Total processing time was 28 secs.
Reproducibility
To reproduce this report, do:
html <- matrixStats:::benchmark('colMeans2')
Copyright Henrik Bengtsson. Last updated on 2021-08-25 22:19:37 (+0200 UTC). Powered by RSP.