matrixStats.benchmarks
binMeans() benchmarks on subsetted computation
This report benchmark the performance of binMeans() on subsetted computation.
Results
Non-sorted simulated data
> nx <- 1e+05
> set.seed(48879)
> x <- runif(nx, min = 0, max = 1)
> y <- runif(nx, min = 0, max = 1)
> nb <- 1000
> bx <- seq(from = 0, to = 1, length.out = nb + 1L)
> bx <- c(-1, bx, 2)
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> y_S <- y[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5166771 276.0 7916910 422.9 7916910 422.9
Vcells 9545628 72.9 33191153 253.3 53339345 407.0
> stats <- microbenchmark(binMeans_x_y_S = binMeans(x = x_S, y = y_S, bx = bx, count = TRUE), `binMeans(x, y, idxs)` = binMeans(x = x,
+ y = y, idxs = idxs, bx = bx, count = TRUE), `binMeans(x[idxs], y[idxs])` = binMeans(x = x[idxs],
+ y = y[idxs], bx = bx, count = TRUE), unit = "ms")
Table: Benchmarking of binMeans_x_y_S(), binMeans(x, y, idxs)() and binMeans(x[idxs], y[idxs])() on unsorted data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
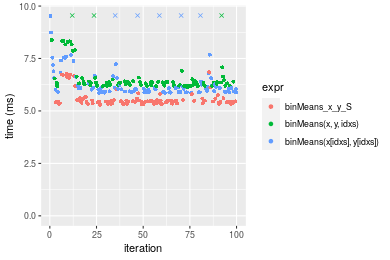
| 1 | binMeans_x_y_S | 5.293469 | 5.367801 | 5.582491 | 5.444456 | 5.526404 | 6.844966 |
| 3 | binMeans(x[idxs], y[idxs]) | 5.859617 | 5.911293 | 6.504632 | 6.054612 | 6.557577 | 10.785069 |
| 2 | binMeans(x, y, idxs) | 6.138208 | 6.209178 | 6.647663 | 6.328808 | 6.417711 | 12.539914 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | binMeans_x_y_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 3 | binMeans(x[idxs], y[idxs]) | 1.106952 | 1.101250 | 1.165184 | 1.112069 | 1.186590 | 1.575621 |
| 2 | binMeans(x, y, idxs) | 1.159581 | 1.156745 | 1.190806 | 1.162432 | 1.161282 | 1.831991 |
Figure: Benchmarking of binMeans_x_y_S(), binMeans(x, y, idxs)() and binMeans(x[idxs], y[idxs])() on unsorted data. Outliers are displayed as crosses. Times are in milliseconds.
Sorted simulated data
> x <- sort(x)
> idxs <- sort(idxs)
> x_S <- x[idxs]
> y_S <- y[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5153160 275.3 7916910 422.9 7916910 422.9
Vcells 9395238 71.7 33191153 253.3 53339345 407.0
> stats <- microbenchmark(binMeans_x_y_S = binMeans(x = x_S, y = y_S, bx = bx, count = TRUE), `binMeans(x, y, idxs)` = binMeans(x = x,
+ y = y, idxs = idxs, bx = bx, count = TRUE), `binMeans(x[idxs], y[idxs])` = binMeans(x = x[idxs],
+ y = y[idxs], bx = bx, count = TRUE), unit = "ms")
Table: Benchmarking of binMeans_x_y_S(), binMeans(x, y, idxs)() and binMeans(x[idxs], y[idxs])() on sorted data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
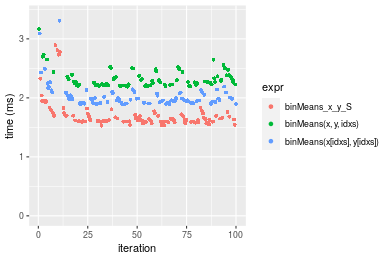
| 1 | binMeans_x_y_S | 1.527029 | 1.598313 | 1.752372 | 1.640097 | 1.726751 | 5.290372 |
| 3 | binMeans(x[idxs], y[idxs]) | 1.891452 | 1.916651 | 2.124493 | 1.980376 | 2.088344 | 5.602020 |
| 2 | binMeans(x, y, idxs) | 2.189744 | 2.223369 | 2.595059 | 2.294065 | 2.485793 | 6.148390 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | binMeans_x_y_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 3 | binMeans(x[idxs], y[idxs]) | 1.238648 | 1.199171 | 1.212353 | 1.207475 | 1.209407 | 1.058909 |
| 2 | binMeans(x, y, idxs) | 1.433990 | 1.391072 | 1.480884 | 1.398737 | 1.439578 | 1.162185 |
Figure: Benchmarking of binMeans_x_y_S(), binMeans(x, y, idxs)() and binMeans(x[idxs], y[idxs])() on sorted data. Outliers are displayed as crosses. Times are in milliseconds.
Appendix
Session information
R version 4.1.1 Patched (2021-08-10 r80727)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.5 LTS
Matrix products: default
BLAS: /home/hb/software/R-devel/R-4-1-branch/lib/R/lib/libRblas.so
LAPACK: /home/hb/software/R-devel/R-4-1-branch/lib/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] microbenchmark_1.4-7 matrixStats_0.60.0 ggplot2_3.3.5
[4] knitr_1.33 R.devices_2.17.0 R.utils_2.10.1
[7] R.oo_1.24.0 R.methodsS3_1.8.1-9001 history_0.0.1-9000
loaded via a namespace (and not attached):
[1] Biobase_2.52.0 httr_1.4.2 splines_4.1.1
[4] bit64_4.0.5 network_1.17.1 assertthat_0.2.1
[7] highr_0.9 stats4_4.1.1 blob_1.2.2
[10] GenomeInfoDbData_1.2.6 robustbase_0.93-8 pillar_1.6.2
[13] RSQLite_2.2.8 lattice_0.20-44 glue_1.4.2
[16] digest_0.6.27 XVector_0.32.0 colorspace_2.0-2
[19] Matrix_1.3-4 XML_3.99-0.7 pkgconfig_2.0.3
[22] zlibbioc_1.38.0 genefilter_1.74.0 purrr_0.3.4
[25] ergm_4.1.2 xtable_1.8-4 scales_1.1.1
[28] tibble_3.1.4 annotate_1.70.0 KEGGREST_1.32.0
[31] farver_2.1.0 generics_0.1.0 IRanges_2.26.0
[34] ellipsis_0.3.2 cachem_1.0.6 withr_2.4.2
[37] BiocGenerics_0.38.0 mime_0.11 survival_3.2-13
[40] magrittr_2.0.1 crayon_1.4.1 statnet.common_4.5.0
[43] memoise_2.0.0 laeken_0.5.1 fansi_0.5.0
[46] R.cache_0.15.0 MASS_7.3-54 R.rsp_0.44.0
[49] progressr_0.8.0 tools_4.1.1 lifecycle_1.0.0
[52] S4Vectors_0.30.0 trust_0.1-8 munsell_0.5.0
[55] tabby_0.0.1-9001 AnnotationDbi_1.54.1 Biostrings_2.60.2
[58] compiler_4.1.1 GenomeInfoDb_1.28.1 rlang_0.4.11
[61] grid_4.1.1 RCurl_1.98-1.4 cwhmisc_6.6
[64] rstudioapi_0.13 rappdirs_0.3.3 startup_0.15.0
[67] labeling_0.4.2 bitops_1.0-7 base64enc_0.1-3
[70] boot_1.3-28 gtable_0.3.0 DBI_1.1.1
[73] markdown_1.1 R6_2.5.1 lpSolveAPI_5.5.2.0-17.7
[76] rle_0.9.2 dplyr_1.0.7 fastmap_1.1.0
[79] bit_4.0.4 utf8_1.2.2 parallel_4.1.1
[82] Rcpp_1.0.7 vctrs_0.3.8 png_0.1-7
[85] DEoptimR_1.0-9 tidyselect_1.1.1 xfun_0.25
[88] coda_0.19-4
Total processing time was 4.21 secs.
Reproducibility
To reproduce this report, do:
html <- matrixStats:::benchmark('binMeans')
Copyright Dongcan Jiang. Last updated on 2021-08-25 22:09:28 (+0200 UTC). Powered by RSP.