matrixStats.benchmarks
binCounts() benchmarks
This report benchmark the performance of binCounts() against alternative methods.
Alternative methods
- hist()
as below
> hist <- graphics::hist
> binCounts_hist <- function(x, bx, right = FALSE, ...) {
+ hist(x, breaks = bx, right = right, include.lowest = TRUE, plot = FALSE)$counts
+ }
Data type “integer”
Non-sorted simulated data
> set.seed(48879)
> nx <- 1e+05
> xmax <- 0.01 * nx
> x <- runif(nx, min = 0, max = xmax)
> storage.mode(x) <- mode
> str(x)
int [1:100000] 722 285 591 3 349 509 216 91 150 383 ...
> nb <- 10000
> bx <- seq(from = 0, to = xmax, length.out = nb + 1L)
> bx <- c(-1, bx, xmax + 1)
Results
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5164682 275.9 7916910 422.9 7916910 422.9
Vcells 9240731 70.6 33191153 253.3 53339345 407.0
> stats <- microbenchmark(binCounts = binCounts(x, bx = bx), hist = binCounts_hist(x, bx = bx), unit = "ms")
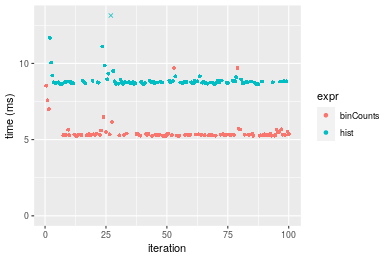
Table: Benchmarking of binCounts() and hist() on integer+unsorted data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | binCounts | 5.221739 | 5.291779 | 5.519676 | 5.322615 | 5.356448 | 9.70044 |
| 2 | hist | 8.632203 | 8.710278 | 8.920086 | 8.757371 | 8.827594 | 14.58375 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | binCounts | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | hist | 1.653128 | 1.646002 | 1.616052 | 1.645314 | 1.648031 | 1.503411 |
Figure: Benchmarking of binCounts() and hist() on integer+unsorted data. Outliers are displayed as crosses. Times are in milliseconds.
Sorted simulated data
> x <- sort(x)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5152060 275.2 7916910 422.9 7916910 422.9
Vcells 9057518 69.2 33191153 253.3 53339345 407.0
> stats <- microbenchmark(binCounts = binCounts(x, bx = bx), hist = binCounts_hist(x, bx = bx), unit = "ms")
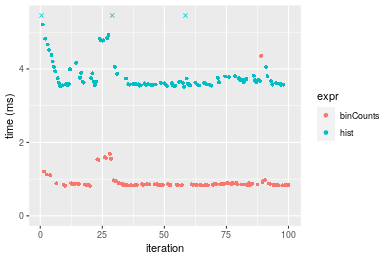
Table: Benchmarking of binCounts() and hist() on integer+sorted data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | binCounts | 0.818483 | 0.838998 | 0.9454563 | 0.8567175 | 0.8816355 | 4.359132 |
| 2 | hist | 3.515065 | 3.572513 | 3.8929789 | 3.6355275 | 3.8108480 | 7.925165 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | binCounts | 1.00000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.00000 |
| 2 | hist | 4.29461 | 4.258071 | 4.117566 | 4.243555 | 4.322476 | 1.81806 |
Figure: Benchmarking of binCounts() and hist() on integer+sorted data. Outliers are displayed as crosses. Times are in milliseconds.
Data type “double”
Non-sorted simulated data
> set.seed(48879)
> nx <- 1e+05
> xmax <- 0.01 * nx
> x <- runif(nx, min = 0, max = xmax)
> storage.mode(x) <- mode
> str(x)
num [1:100000] 722.11 285.54 591.33 3.42 349.14 ...
> nb <- 10000
> bx <- seq(from = 0, to = xmax, length.out = nb + 1L)
> bx <- c(-1, bx, xmax + 1)
Results
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5152123 275.2 7916910 422.9 7916910 422.9
Vcells 9107560 69.5 33191153 253.3 53339345 407.0
> stats <- microbenchmark(binCounts = binCounts(x, bx = bx), hist = binCounts_hist(x, bx = bx), unit = "ms")
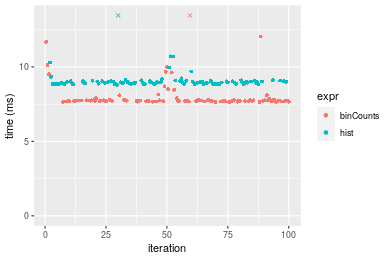
Table: Benchmarking of binCounts() and hist() on double+unsorted data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | binCounts | 7.638411 | 7.690463 | 8.003047 | 7.732960 | 7.758226 | 13.53363 |
| 2 | hist | 8.804883 | 8.872464 | 9.066958 | 8.953775 | 9.030924 | 13.77090 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | binCounts | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | hist | 1.152711 | 1.153697 | 1.132938 | 1.157872 | 1.164045 | 1.017532 |
Figure: Benchmarking of binCounts() and hist() on double+unsorted data. Outliers are displayed as crosses. Times are in milliseconds.
Sorted simulated data
> x <- sort(x)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5152189 275.2 7916910 422.9 7916910 422.9
Vcells 9108116 69.5 33191153 253.3 53339345 407.0
> stats <- microbenchmark(binCounts = binCounts(x, bx = bx), hist = binCounts_hist(x, bx = bx), unit = "ms")
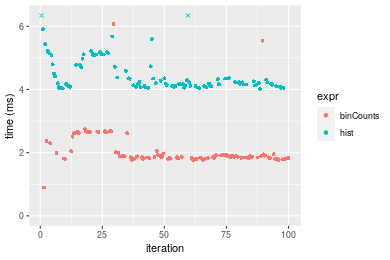
Table: Benchmarking of binCounts() and hist() on double+sorted data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | binCounts | 0.899685 | 1.82550 | 2.075927 | 1.878453 | 1.991132 | 6.068830 |
| 2 | hist | 4.017487 | 4.11005 | 4.483815 | 4.206721 | 4.750952 | 8.044012 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | binCounts | 1.000000 | 1.000000 | 1.000000 | 1.00000 | 1.000000 | 1.000000 |
| 2 | hist | 4.465437 | 2.251465 | 2.159909 | 2.23946 | 2.386056 | 1.325463 |
Figure: Benchmarking of binCounts() and hist() on double+sorted data. Outliers are displayed as crosses. Times are in milliseconds.
Appendix
Session information
R version 4.1.1 Patched (2021-08-10 r80727)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.5 LTS
Matrix products: default
BLAS: /home/hb/software/R-devel/R-4-1-branch/lib/R/lib/libRblas.so
LAPACK: /home/hb/software/R-devel/R-4-1-branch/lib/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] microbenchmark_1.4-7 matrixStats_0.60.0 ggplot2_3.3.5
[4] knitr_1.33 R.devices_2.17.0 R.utils_2.10.1
[7] R.oo_1.24.0 R.methodsS3_1.8.1-9001 history_0.0.1-9000
loaded via a namespace (and not attached):
[1] Biobase_2.52.0 httr_1.4.2 splines_4.1.1
[4] bit64_4.0.5 network_1.17.1 assertthat_0.2.1
[7] highr_0.9 stats4_4.1.1 blob_1.2.2
[10] GenomeInfoDbData_1.2.6 robustbase_0.93-8 pillar_1.6.2
[13] RSQLite_2.2.8 lattice_0.20-44 glue_1.4.2
[16] digest_0.6.27 XVector_0.32.0 colorspace_2.0-2
[19] Matrix_1.3-4 XML_3.99-0.7 pkgconfig_2.0.3
[22] zlibbioc_1.38.0 genefilter_1.74.0 purrr_0.3.4
[25] ergm_4.1.2 xtable_1.8-4 scales_1.1.1
[28] tibble_3.1.4 annotate_1.70.0 KEGGREST_1.32.0
[31] farver_2.1.0 generics_0.1.0 IRanges_2.26.0
[34] ellipsis_0.3.2 cachem_1.0.6 withr_2.4.2
[37] BiocGenerics_0.38.0 mime_0.11 survival_3.2-13
[40] magrittr_2.0.1 crayon_1.4.1 statnet.common_4.5.0
[43] memoise_2.0.0 laeken_0.5.1 fansi_0.5.0
[46] R.cache_0.15.0 MASS_7.3-54 R.rsp_0.44.0
[49] progressr_0.8.0 tools_4.1.1 lifecycle_1.0.0
[52] S4Vectors_0.30.0 trust_0.1-8 munsell_0.5.0
[55] tabby_0.0.1-9001 AnnotationDbi_1.54.1 Biostrings_2.60.2
[58] compiler_4.1.1 GenomeInfoDb_1.28.1 rlang_0.4.11
[61] grid_4.1.1 RCurl_1.98-1.4 cwhmisc_6.6
[64] rstudioapi_0.13 rappdirs_0.3.3 startup_0.15.0
[67] labeling_0.4.2 bitops_1.0-7 base64enc_0.1-3
[70] boot_1.3-28 gtable_0.3.0 DBI_1.1.1
[73] markdown_1.1 R6_2.5.1 lpSolveAPI_5.5.2.0-17.7
[76] rle_0.9.2 dplyr_1.0.7 fastmap_1.1.0
[79] bit_4.0.4 utf8_1.2.2 parallel_4.1.1
[82] Rcpp_1.0.7 vctrs_0.3.8 png_0.1-7
[85] DEoptimR_1.0-9 tidyselect_1.1.1 xfun_0.25
[88] coda_0.19-4
Total processing time was 7.7 secs.
Reproducibility
To reproduce this report, do:
html <- matrixStats:::benchmark('binCounts')
Copyright Henrik Bengtsson. Last updated on 2021-08-25 22:09:23 (+0200 UTC). Powered by RSP.