matrixStats.benchmarks
anyMissing() benchmarks on subsetted computation
This report benchmark the performance of anyMissing() on subsetted computation.
Data type “integer”
Data
> rvector <- function(n, mode = c("logical", "double", "integer"), range = c(-100, +100), na_prob = 0) {
+ mode <- match.arg(mode)
+ if (mode == "logical") {
+ x <- sample(c(FALSE, TRUE), size = n, replace = TRUE)
+ } else {
+ x <- runif(n, min = range[1], max = range[2])
+ }
+ storage.mode(x) <- mode
+ if (na_prob > 0)
+ x[sample(n, size = na_prob * n)] <- NA
+ x
+ }
> rvectors <- function(scale = 10, seed = 1, ...) {
+ set.seed(seed)
+ data <- list()
+ data[[1]] <- rvector(n = scale * 100, ...)
+ data[[2]] <- rvector(n = scale * 1000, ...)
+ data[[3]] <- rvector(n = scale * 10000, ...)
+ data[[4]] <- rvector(n = scale * 1e+05, ...)
+ data[[5]] <- rvector(n = scale * 1e+06, ...)
+ names(data) <- sprintf("n = %d", sapply(data, FUN = length))
+ data
+ }
> data <- rvectors(mode = mode)
Results
n = 1000 vector
> x <- data[["n = 1000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5162916 275.8 7916910 422.9 7916910 422.9
Vcells 25710602 196.2 53955392 411.7 50098692 382.3
> stats <- microbenchmark(anyMissing_x_S = anyMissing(x_S), `anyMissing(x, idxs)` = anyMissing(x, idxs = idxs),
+ `anyMissing(x[idxs])` = anyMissing(x[idxs]), unit = "ms")
Table: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on integer+n = 1000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 0.001299 | 0.0013310 | 0.0014001 | 0.0013850 | 0.0014175 | 0.002203 |
| 2 | anyMissing(x, idxs) | 0.002861 | 0.0029085 | 0.0029626 | 0.0029380 | 0.0029755 | 0.004156 |
| 3 | anyMissing(x[idxs]) | 0.003853 | 0.0039675 | 0.0052384 | 0.0040435 | 0.0041210 | 0.121189 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | anyMissing(x, idxs) | 2.202463 | 2.185199 | 2.116021 | 2.121300 | 2.099118 | 1.886518 |
| 3 | anyMissing(x[idxs]) | 2.966128 | 2.980841 | 3.741459 | 2.919495 | 2.907231 | 55.010894 |
Figure: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on integer+n = 1000 data. Outliers are displayed as crosses. Times are in milliseconds.
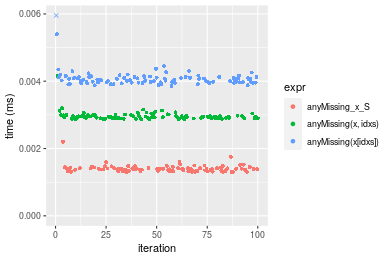
n = 10000 vector
> x <- data[["n = 10000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5148112 275 7916910 422.9 7916910 422.9
Vcells 14536728 111 53955392 411.7 50098692 382.3
> stats <- microbenchmark(anyMissing_x_S = anyMissing(x_S), `anyMissing(x, idxs)` = anyMissing(x, idxs = idxs),
+ `anyMissing(x[idxs])` = anyMissing(x[idxs]), unit = "ms")
Table: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on integer+n = 10000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 0.006821 | 0.006938 | 0.0071182 | 0.0070145 | 0.0071065 | 0.012310 |
| 2 | anyMissing(x, idxs) | 0.021872 | 0.021980 | 0.0226106 | 0.0220715 | 0.0222115 | 0.037837 |
| 3 | anyMissing(x[idxs]) | 0.028426 | 0.028889 | 0.0302136 | 0.0292280 | 0.0297295 | 0.067615 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 1.000000 | 1.00000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | anyMissing(x, idxs) | 3.206568 | 3.16806 | 3.176452 | 3.146554 | 3.125519 | 3.073680 |
| 3 | anyMissing(x[idxs]) | 4.167424 | 4.16388 | 4.244565 | 4.166797 | 4.183424 | 5.492689 |
Figure: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on integer+n = 10000 data. Outliers are displayed as crosses. Times are in milliseconds.
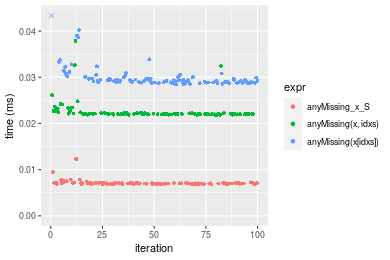
n = 100000 vector
> x <- data[["n = 100000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5148184 275.0 7916910 422.9 7916910 422.9
Vcells 14600288 111.4 53955392 411.7 50098692 382.3
> stats <- microbenchmark(anyMissing_x_S = anyMissing(x_S), `anyMissing(x, idxs)` = anyMissing(x, idxs = idxs),
+ `anyMissing(x[idxs])` = anyMissing(x[idxs]), unit = "ms")
Table: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on integer+n = 100000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 0.045269 | 0.047030 | 0.0565744 | 0.055161 | 0.0605690 | 0.097646 |
| 2 | anyMissing(x, idxs) | 0.189848 | 0.196721 | 0.2237074 | 0.210139 | 0.2433385 | 0.385652 |
| 3 | anyMissing(x[idxs]) | 0.222029 | 0.238814 | 0.2838194 | 0.268992 | 0.2986360 | 0.765679 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | anyMissing(x, idxs) | 4.193775 | 4.182883 | 3.954212 | 3.809557 | 4.017542 | 3.949491 |
| 3 | anyMissing(x[idxs]) | 4.904659 | 5.077908 | 5.016743 | 4.876489 | 4.930509 | 7.841376 |
Figure: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on integer+n = 100000 data. Outliers are displayed as crosses. Times are in milliseconds.
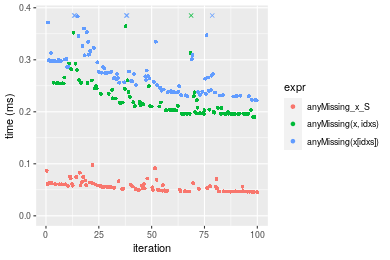
n = 1000000 vector
> x <- data[["n = 1000000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5148256 275.0 7916910 422.9 7916910 422.9
Vcells 15230337 116.2 53955392 411.7 50098692 382.3
> stats <- microbenchmark(anyMissing_x_S = anyMissing(x_S), `anyMissing(x, idxs)` = anyMissing(x, idxs = idxs),
+ `anyMissing(x[idxs])` = anyMissing(x[idxs]), unit = "ms")
Table: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on integer+n = 1000000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 0.454448 | 0.5216525 | 0.5786677 | 0.5436835 | 0.5899715 | 1.079747 |
| 2 | anyMissing(x, idxs) | 2.487519 | 2.7614475 | 3.4705498 | 3.3085560 | 3.8648375 | 7.685493 |
| 3 | anyMissing(x[idxs]) | 3.827613 | 4.4859705 | 5.4221502 | 4.9909670 | 5.9684330 | 10.399041 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | anyMissing(x, idxs) | 5.473715 | 5.293653 | 5.997483 | 6.085445 | 6.550889 | 7.117865 |
| 3 | anyMissing(x[idxs]) | 8.422554 | 8.599538 | 9.370058 | 9.179913 | 10.116477 | 9.630998 |
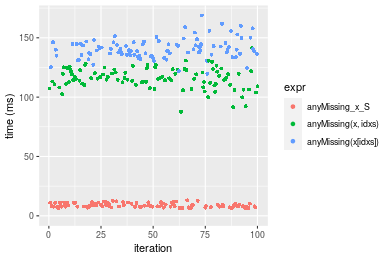
Figure: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on integer+n = 1000000 data. Outliers are displayed as crosses. Times are in milliseconds.
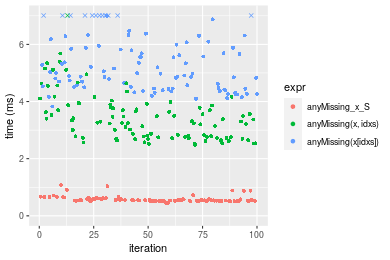
n = 10000000 vector
> x <- data[["n = 10000000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5148328 275.0 7916910 422.9 7916910 422.9
Vcells 21530590 164.3 53955392 411.7 50658010 386.5
> stats <- microbenchmark(anyMissing_x_S = anyMissing(x_S), `anyMissing(x, idxs)` = anyMissing(x, idxs = idxs),
+ `anyMissing(x[idxs])` = anyMissing(x[idxs]), unit = "ms")
Table: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on integer+n = 10000000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 6.390179 | 7.663246 | 8.921722 | 8.455134 | 10.36377 | 13.00473 |
| 2 | anyMissing(x, idxs) | 87.748499 | 111.039786 | 115.822160 | 115.685753 | 122.27734 | 141.40928 |
| 3 | anyMissing(x[idxs]) | 119.607946 | 135.674981 | 140.726406 | 139.743561 | 144.71224 | 168.76377 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 1.00000 | 1.00000 | 1.00000 | 1.00000 | 1.00000 | 1.00000 |
| 2 | anyMissing(x, idxs) | 13.73177 | 14.48992 | 12.98204 | 13.68231 | 11.79853 | 10.87368 |
| 3 | anyMissing(x[idxs]) | 18.71746 | 17.70464 | 15.77346 | 16.52766 | 13.96328 | 12.97711 |
Figure: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on integer+n = 10000000 data. Outliers are displayed as crosses. Times are in milliseconds.
Data type “double”
Data
> rvector <- function(n, mode = c("logical", "double", "integer"), range = c(-100, +100), na_prob = 0) {
+ mode <- match.arg(mode)
+ if (mode == "logical") {
+ x <- sample(c(FALSE, TRUE), size = n, replace = TRUE)
+ } else {
+ x <- runif(n, min = range[1], max = range[2])
+ }
+ storage.mode(x) <- mode
+ if (na_prob > 0)
+ x[sample(n, size = na_prob * n)] <- NA
+ x
+ }
> rvectors <- function(scale = 10, seed = 1, ...) {
+ set.seed(seed)
+ data <- list()
+ data[[1]] <- rvector(n = scale * 100, ...)
+ data[[2]] <- rvector(n = scale * 1000, ...)
+ data[[3]] <- rvector(n = scale * 10000, ...)
+ data[[4]] <- rvector(n = scale * 1e+05, ...)
+ data[[5]] <- rvector(n = scale * 1e+06, ...)
+ names(data) <- sprintf("n = %d", sapply(data, FUN = length))
+ data
+ }
> data <- rvectors(mode = mode)
Results
n = 1000 vector
> x <- data[["n = 1000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5148409 275.0 7916910 422.9 7916910 422.9
Vcells 20087457 153.3 53955392 411.7 53032641 404.7
> stats <- microbenchmark(anyMissing_x_S = anyMissing(x_S), `anyMissing(x, idxs)` = anyMissing(x, idxs = idxs),
+ `anyMissing(x[idxs])` = anyMissing(x[idxs]), unit = "ms")
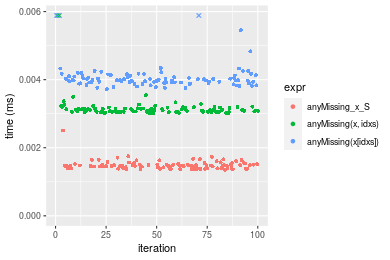
Table: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on double+n = 1000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 0.001356 | 0.0014020 | 0.0014909 | 0.0014710 | 0.0015125 | 0.002507 |
| 2 | anyMissing(x, idxs) | 0.003008 | 0.0030575 | 0.0032146 | 0.0030975 | 0.0031555 | 0.009546 |
| 3 | anyMissing(x[idxs]) | 0.003724 | 0.0039190 | 0.0047963 | 0.0039910 | 0.0041330 | 0.049398 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 1.000000 | 1.000000 | 1.000000 | 1.00000 | 1.000000 | 1.000000 |
| 2 | anyMissing(x, idxs) | 2.218289 | 2.180813 | 2.156126 | 2.10571 | 2.086281 | 3.807738 |
| 3 | anyMissing(x[idxs]) | 2.746313 | 2.795292 | 3.217042 | 2.71312 | 2.732562 | 19.704029 |
Figure: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on double+n = 1000 data. Outliers are displayed as crosses. Times are in milliseconds.
n = 10000 vector
> x <- data[["n = 10000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5148472 275.0 7916910 422.9 7916910 422.9
Vcells 20097246 153.4 53955392 411.7 53032641 404.7
> stats <- microbenchmark(anyMissing_x_S = anyMissing(x_S), `anyMissing(x, idxs)` = anyMissing(x, idxs = idxs),
+ `anyMissing(x[idxs])` = anyMissing(x[idxs]), unit = "ms")
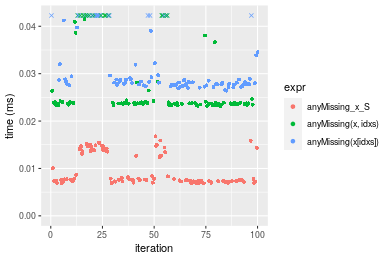
Table: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on double+n = 10000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 0.006850 | 0.0072490 | 0.0094397 | 0.0075730 | 0.0132905 | 0.016769 |
| 2 | anyMissing(x, idxs) | 0.023116 | 0.0235495 | 0.0286511 | 0.0238020 | 0.0282000 | 0.049185 |
| 3 | anyMissing(x[idxs]) | 0.026394 | 0.0276825 | 0.0337686 | 0.0282005 | 0.0330330 | 0.065500 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | anyMissing(x, idxs) | 3.374598 | 3.248655 | 3.035167 | 3.143008 | 2.121816 | 2.933091 |
| 3 | anyMissing(x[idxs]) | 3.853139 | 3.818803 | 3.577295 | 3.723822 | 2.485460 | 3.906017 |
Figure: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on double+n = 10000 data. Outliers are displayed as crosses. Times are in milliseconds.
n = 100000 vector
> x <- data[["n = 100000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5148544 275.0 7916910 422.9 7916910 422.9
Vcells 20192161 154.1 53955392 411.7 53032641 404.7
> stats <- microbenchmark(anyMissing_x_S = anyMissing(x_S), `anyMissing(x, idxs)` = anyMissing(x, idxs = idxs),
+ `anyMissing(x[idxs])` = anyMissing(x[idxs]), unit = "ms")
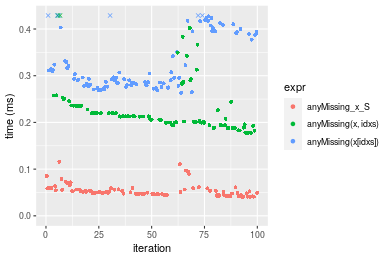
Table: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on double+n = 100000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 0.040801 | 0.0458760 | 0.0535020 | 0.0495030 | 0.0560180 | 0.115687 |
| 2 | anyMissing(x, idxs) | 0.177440 | 0.1994325 | 0.2273533 | 0.2125495 | 0.2316585 | 0.549579 |
| 3 | anyMissing(x[idxs]) | 0.263107 | 0.2803375 | 0.3378248 | 0.2959000 | 0.3937720 | 1.195971 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 2 | anyMissing(x, idxs) | 4.348913 | 4.347208 | 4.249433 | 4.293669 | 4.135430 | 4.750568 |
| 3 | anyMissing(x[idxs]) | 6.448543 | 6.110766 | 6.314242 | 5.977416 | 7.029383 | 10.337990 |
Figure: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on double+n = 100000 data. Outliers are displayed as crosses. Times are in milliseconds.
n = 1000000 vector
> x <- data[["n = 1000000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5148616 275.0 7916910 422.9 7916910 422.9
Vcells 21137210 161.3 53955392 411.7 53032641 404.7
> stats <- microbenchmark(anyMissing_x_S = anyMissing(x_S), `anyMissing(x, idxs)` = anyMissing(x, idxs = idxs),
+ `anyMissing(x[idxs])` = anyMissing(x[idxs]), unit = "ms")
Table: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on double+n = 1000000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 0.635010 | 0.746746 | 0.7843915 | 0.762187 | 0.786418 | 1.741441 |
| 2 | anyMissing(x, idxs) | 5.124992 | 5.398494 | 6.3265125 | 5.801729 | 6.654898 | 11.951174 |
| 3 | anyMissing(x[idxs]) | 6.766017 | 9.684119 | 10.3313381 | 10.067975 | 10.829647 | 22.629365 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 1.000000 | 1.000000 | 1.000000 | 1.00000 | 1.000000 | 1.000000 |
| 2 | anyMissing(x, idxs) | 8.070726 | 7.229358 | 8.065504 | 7.61195 | 8.462292 | 6.862807 |
| 3 | anyMissing(x[idxs]) | 10.654977 | 12.968425 | 13.171150 | 13.20932 | 13.770853 | 12.994620 |
Figure: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on double+n = 1000000 data. Outliers are displayed as crosses. Times are in milliseconds.
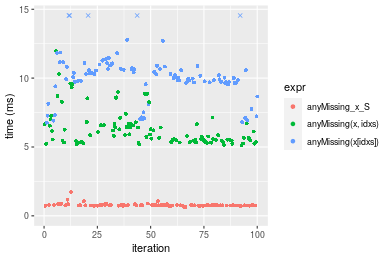
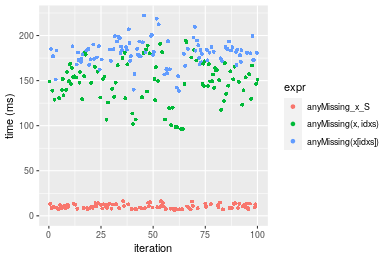
n = 10000000 vector
> x <- data[["n = 10000000"]]
> idxs <- sample.int(length(x), size = length(x) * 0.7)
> x_S <- x[idxs]
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 5148688 275.0 7916910 422.9 7916910 422.9
Vcells 30587693 233.4 53955392 411.7 53339345 407.0
> stats <- microbenchmark(anyMissing_x_S = anyMissing(x_S), `anyMissing(x, idxs)` = anyMissing(x, idxs = idxs),
+ `anyMissing(x[idxs])` = anyMissing(x[idxs]), unit = "ms")
Table: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on double+n = 10000000 data. The top panel shows times in milliseconds and the bottom panel shows relative times.
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 6.503693 | 8.108895 | 10.21231 | 9.51436 | 12.53715 | 16.24656 |
| 2 | anyMissing(x, idxs) | 96.152599 | 132.862656 | 148.87533 | 149.98104 | 165.32895 | 194.07407 |
| 3 | anyMissing(x[idxs]) | 138.857610 | 172.913414 | 180.78766 | 181.79698 | 187.64958 | 222.23205 |
| expr | min | lq | mean | median | uq | max | |
|---|---|---|---|---|---|---|---|
| 1 | anyMissing_x_S | 1.00000 | 1.00000 | 1.00000 | 1.00000 | 1.00000 | 1.00000 |
| 2 | anyMissing(x, idxs) | 14.78431 | 16.38480 | 14.57803 | 15.76365 | 13.18712 | 11.94555 |
| 3 | anyMissing(x[idxs]) | 21.35058 | 21.32392 | 17.70292 | 19.10764 | 14.96748 | 13.67872 |
Figure: Benchmarking of anyMissing_x_S(), anyMissing(x, idxs)() and anyMissing(x[idxs])() on double+n = 10000000 data. Outliers are displayed as crosses. Times are in milliseconds.
Appendix
Session information
R version 4.1.1 Patched (2021-08-10 r80727)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.5 LTS
Matrix products: default
BLAS: /home/hb/software/R-devel/R-4-1-branch/lib/R/lib/libRblas.so
LAPACK: /home/hb/software/R-devel/R-4-1-branch/lib/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] microbenchmark_1.4-7 matrixStats_0.60.0 ggplot2_3.3.5
[4] knitr_1.33 R.devices_2.17.0 R.utils_2.10.1
[7] R.oo_1.24.0 R.methodsS3_1.8.1-9001 history_0.0.1-9000
loaded via a namespace (and not attached):
[1] Biobase_2.52.0 httr_1.4.2 splines_4.1.1
[4] bit64_4.0.5 network_1.17.1 assertthat_0.2.1
[7] highr_0.9 stats4_4.1.1 blob_1.2.2
[10] GenomeInfoDbData_1.2.6 robustbase_0.93-8 pillar_1.6.2
[13] RSQLite_2.2.8 lattice_0.20-44 glue_1.4.2
[16] digest_0.6.27 XVector_0.32.0 colorspace_2.0-2
[19] Matrix_1.3-4 XML_3.99-0.7 pkgconfig_2.0.3
[22] zlibbioc_1.38.0 genefilter_1.74.0 purrr_0.3.4
[25] ergm_4.1.2 xtable_1.8-4 scales_1.1.1
[28] tibble_3.1.4 annotate_1.70.0 KEGGREST_1.32.0
[31] farver_2.1.0 generics_0.1.0 IRanges_2.26.0
[34] ellipsis_0.3.2 cachem_1.0.6 withr_2.4.2
[37] BiocGenerics_0.38.0 mime_0.11 survival_3.2-13
[40] magrittr_2.0.1 crayon_1.4.1 statnet.common_4.5.0
[43] memoise_2.0.0 laeken_0.5.1 fansi_0.5.0
[46] R.cache_0.15.0 MASS_7.3-54 R.rsp_0.44.0
[49] progressr_0.8.0 tools_4.1.1 lifecycle_1.0.0
[52] S4Vectors_0.30.0 trust_0.1-8 munsell_0.5.0
[55] tabby_0.0.1-9001 AnnotationDbi_1.54.1 Biostrings_2.60.2
[58] compiler_4.1.1 GenomeInfoDb_1.28.1 rlang_0.4.11
[61] grid_4.1.1 RCurl_1.98-1.4 cwhmisc_6.6
[64] rstudioapi_0.13 rappdirs_0.3.3 startup_0.15.0
[67] labeling_0.4.2 bitops_1.0-7 base64enc_0.1-3
[70] boot_1.3-28 gtable_0.3.0 DBI_1.1.1
[73] markdown_1.1 R6_2.5.1 lpSolveAPI_5.5.2.0-17.7
[76] rle_0.9.2 dplyr_1.0.7 fastmap_1.1.0
[79] bit_4.0.4 utf8_1.2.2 parallel_4.1.1
[82] Rcpp_1.0.7 vctrs_0.3.8 png_0.1-7
[85] DEoptimR_1.0-9 tidyselect_1.1.1 xfun_0.25
[88] coda_0.19-4
Total processing time was 1.26 mins.
Reproducibility
To reproduce this report, do:
html <- matrixStats:::benchmark('anyMissing_subset')
Copyright Dongcan Jiang. Last updated on 2021-08-25 22:08:46 (+0200 UTC). Powered by RSP.